Cucurbit Genetics Cooperative Report 12:46-49 (article 20) 1989
Raffo, A.J, I.A. Khan, L.F. Lippert, M.O. Hall, and G.E. Jones
Department of Plant Pathology, University of California, Riverside, CA 92521 (first author); Department of Horticulture, University of Agriculture, Faisalabad, Pakistan (second author); Department of Botany and Plant Sciences, University of California, Riverside, CA 92521, USA (third, fourth, and fifth authors)
Breeding for disease resistance is often restricted due to the lack of reliable and efficient screening procedures. Resistant individuals may be identified by symptomatology, bioassay, and/or serology; however, in some situations, these methods are of limited applicability. Zucchini yellow mosaic virus (ZYMV), a recently reported virus in the potyvirus group ($), has caused appreciable economic losses in many cucurbit species. The economic impact of this virus has been especially significant on various types of melons growing in the irrigated semi-arid Imperial Valley of California. Melon line PI 414723 has been identified as a source of dominant, single gene resistance to this virus (7). Our group has been utilizing this material in a breeding program designed to transfer ZYMV resistance into the western shipping-type muskmelon. We have previously reported a procedure to vegetatively propagate the breeding progenies, as well as a two-step evaluation of this material (3). In this report, we describe a procedure based on the use of cDNA probes to screen for ZYMV resistance in our laboratory. Similar procedures have been developed for the identification of other viruses and viroids and have been found to be highly sensitive and efficient (1, 6, 8.).
cDNA copies of several regions of the ZYMV genome were developed by AJR according to standard procedures of cDNA cloning (5). The cDNA clones were tested for their specific homology to the ZYMV genomic RNA and not to several closely related potyviruses by hybridization (DNA/RNA) before using them as diagnostic probes. Out of approximately two hundred clones tested, several dozen had a high degree of homology and selectivity for our ZYMV isolate. The sensitivity was found to be in the picogram range of viral RNA.
For screening purposes, the breeding progenies were vegetatively propagated (3). The plants were mechanically inoculated with ZYMV freshly extracted by grinding leaves of a source squash plant (‘Early Prolific’ zucchini) in a 10 mM Potassium Phosphate buffer, pH 7.0 with 1% (w/v) celite as an abrasive. It has been observed that the virus infects most efficaciously under greenhouse conditions if the plants are infected at the 2-3 new leaf stage after being repropagated. The systemic mosaic symptoms appear within 2-3 weeks post-inoculation.
Leaves of these plants are harvested (1-5 gm of tissue) and ground in liquid nitrogen. The frozen powdered tissue is soaked in 12 ml of a 1 X SET solution [1% SDS (sodium dodecyl sulfate), 1 mM EDTA, 25 mM TRS-HCL pH 7.5], and 0.5 ml of 10 mg/ml Protease K for approximately 2 hr at 37°C. The extract is centrifuged at 10,000 g for 15 min and the pellet discarded. The supernatant is treated with 0.5 ml of 10 M ammonium acetate and 25 ml of 95% ethanol and the nucleic acids allowed to precipitate at -20°C. The pellet is collected by centrifugation, as described above, air dried, and redissolved in 1X SET and reprecipitated. This is repeated 2-3 times until its spectroscopic analysis showed it to be fairly pure nucleic acid (260/280 ratio greater than 1.7). Finally, the pale green pellet is resuspended in 0.5 ml of 1X SET and the nucleic acid content measured by spectroscopy. All samples are first adjusted for the same amount of nucleic acid, then denatured with 7.5% formaldehyde at 65°C for 10-15 min, brought to 10X SSC (1.5 M Sodium Chloride and 150 mM Sodium Citrate pH 7.0) and finally spotted under vacuum onto nitrocellulose paper in a 96-well manifold (Bethesda Research Laboratories). The nitrocellulose paper is baked at 80°C for 90 min to fix the nucleic acids onto this paper. The unbound portion of the nitrocellulose paper is blocked by prehybridization at 65°C overnight with 100 µg/ml denatured salmon sperm DNA and 5X Denhardt’s solution (1% polyvinylpyrolidone, 1% ficoll, and 1% Bovine Serum Albumin) in 0.5% SDS and 6 X SSC. Hybridization to the nicktranslated (P32labelled according to 5) cDNA probe of ZYMV is carried out in the same solution, this time with probe at 65°C, now for approximately 2 days. Unhybridized probe is stringently washed off the paper prior to autoradiography; first 2 times in 2X SCC and 0.1% SDS at 65°C for at least 30 min each 2 then additional times in 0.1X SSC and 0.1% SDS at 65 ˚ C, for similar times.
The data presented in Fig. 1 is a representative example. Lanes 1-12 are as follows: 1-3 are uninfected healthy controls; 4, 5, and 6 are inoculated F2 plants without symptoms; 7 is a ‘Top Mark’ plant with symptoms; 8-11 are field samples showing some type of mosaic symptoms and lane 12 is a squash plant infected with ZYMV as a positive control. The presence of spots at lanes 4, 6, 7, and 12 and absence at 1-3 show that this probe is capable of hybridizing to ZYMV. The absence at lane 5 reflects resistance in this F2 segregate, while ZYMV detection in lanes 4 and 6 may suggest tolerance in these F2 segregates. The lack of detection at lanes 8-11 indicates that this probe does not hybridize to false positives. These field samples had mosaic symptoms apparently from an infection by another virus and not from ZYMV. Samples from other field plants, not connected with this study, but infected with ZYMV have tested positive with this probe (AJR, personal communication).
The relative intensities of the dots shown in Figure 1 were quantified using a LKB Ultroscan XL laser densitometer. These values are presented in Table 1 as the area of the dot’s peak adjusted for 1 μg of total nucleic acid applied. These figures reflect the conclusions discussed above while allowing for a comparison of the relative titer of virus in each sample. The virus levels in the symptomless F2 plants seen in lanes 4 and 6 are approximately 30% of the levels found in the susceptible plant presented in lane 7.
the resistance to ZYMV has been considered as a Mendelian character. The data presented by Pitrat (7), based on symptomatology, can be explained by a single dominant gene (Zym) and is similar to Tobacco Mosaic Virus resistance found in Nicotiana glutinosa (conferred by the N gene) (2). While preliminary, our data suggest a more complex genetics. The finding of tolerance, as well as the ability to easily sort infected F2 plants in the greenhouse, on the basis of severity of symptoms, into several classes is similar to Ryegrass Mosaic Virus resistance in rye grass (9). This resistance is considered polygenic.
The authors again wish to stress the preliminary nature of our data, however, we also wish to stress the power of the molecular probe. Not only can the cDNA probe be used to detect and quantitate a specific viral presence, and thus be useful in a breeding program such as ours, but the molecular probe can be used to help elucidate the genetics of resistance as well as to identify a possible source of viral potential in an otherwise healthy population.
Figure 1. Dot blot analysis of plant extracts hybridized to a radiolabelled cDNA probe to ZYMV isolated from the Imperial Valley of California. Pleas see text for lane designations. μg refers to the amount of total nucleic acids applied to each dot in that row.
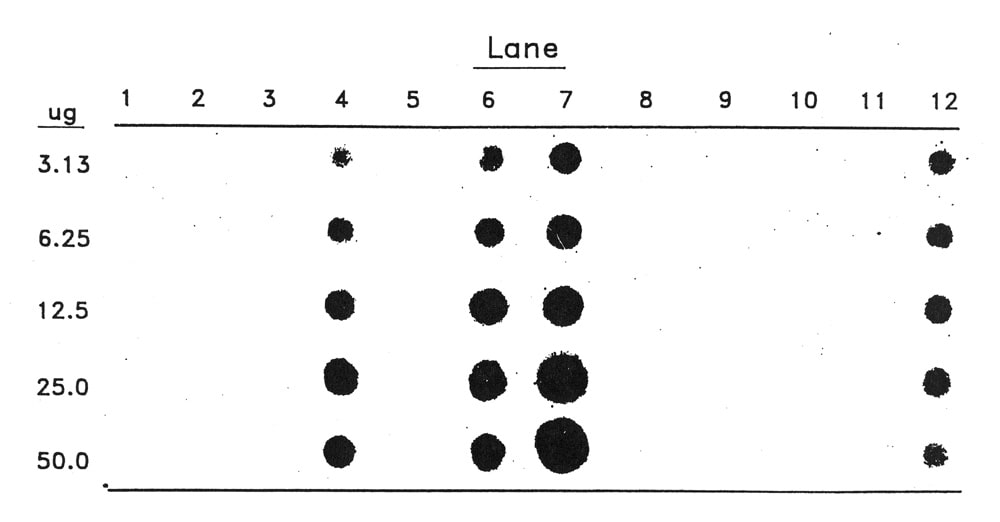
Table 1. Relative quantitation of ZYMV levels from dot blot analysis presented in Figure 1.
Lane z |
Rel. amount y |
Lane z |
Rel. amount y |
| 1 | 0.000 | 7 | 0.774 + 0.084 |
| 2 | 0.000 | 8 | 0.000 |
| 3 | 0.000 | 9 | 0.000 |
| 4 | 0.249 + 0.019 | 10 | 0.000 |
| 5 | 0.000 | 11 | 0.000 |
| 6 | 0.215 + 0.006 | 12 | 0.110 + 0.015 |
z Please see text for lane designations.
y Relative amounts are in absorbance X peak width (mm) adjusted for one microgram of total nucleic acid.
Literature Cited
- Candresse, T., G. Macquaire, M. Monsion, and J. Dunez. 1988. Detection of chrysanthemum stunt viroid (CSV) using nick-translated probes in a dot-blot hybridization assay. J. Virological Methods 20:185-193.
- Holmes, F.O. 1938. Inheritance of resistance to tobacco mosaic disease in tobacco. Phytopathology 28:553-561.
- Khan, I.A., L.F. Lippert, M.O. Hall, and G.E. Jones. 1988. A simple procedures and the genetic potential for rooting of stem cuttings in muskmelon cucurbit. Genet. Coop. Rept. 11:43-46.
- Lisa, V., G. Boccardo,.G. D’agostino, G. Dellavalle, and M. d’Aquillo. 1981. Characterization of a potyvirus that causes zucchini yellow mosaic. Phytopathology 71:667-672.
- Maniatis, T., E.F. Fritsch, and J, Sambrook. 1982. “Molecular cloning; a Laboratory Manual.” Cold Springs Harbor Laboratory.
- Maule, A.J., R. Hull and J. Donson. 1983. The application of spot hybridization to the detection of DNA and RNA viruses in plant tissues. J. Virological Methods 6:215-224.
- Pitrat, M. and H. Lecoq. 1984. Inheritance of zucchini yellow mosaic virus resistance in Cucumis melo L. Euphytica 33:57-61.
- Salager, L.F., R.A. Owens, D.R. Smith and T.O. Diener. 1983. Detection of potato spindle tuber viroid by nucleic acid spot hybridization: evaluation with potato spindle tuber viroid by nucleic acid spot hybridization: evaluation with tuber sprouts and true potato seed. Amer. Potato J. 60:587-597.
- Willkins, P.W. 1974. Tolerance to ryegrass mosaic virus and its inheritance. Ann. Appl. Biol. 78:187-192.