Cucurbit Genetics Cooperative Report 18:31-33 (article 14) 1995
S. Matsuura
Tohoku Seed Company, 1625 Himuro, Nishihara, Utsunomiya 321-32, Japan
Linkage analysis has been performed in cucumber using RFLP markers and the acr locus (5). At that time, almost all of the RFLP clones segregated in Mendelian ratios, except for two genomic clones (B-174 and P-146) which resulted in F2 progeny with the same RFLP pattern as that of paternal lines.
Since polymorphisms were observed between the two inbred lines homozygous for these clones, monomorphic banding patterns were not predicted. Therefore, we studied the genotypes of F1 plants and origin of these two clones to determine the genetic nature of this phenomenon.
Methods. Ten adapted inbred lines and one wild strain were reciprocally crossed to produce F1 progeny. To assure the genetic purity of these parental lines, samples of total DNA for Southern hybridization were isolated from parental germplasm. Mitochondrial and chloroplast DNA was isolated from F1 progeny. To assure the genetic purity of these parental lines, samples of total DNA for Southern hybridization were isolated from parental germplasm,. Mitochondrial and chloroplast DNA was isolated from F1 progeny of a cross between NB-1 and GF-1. Experimental procedures to isolate total, mitochondrial and chloroplast DNAs were according to Murray and Thompson (7), Umbeck and Gengenback (9), and Hirai et al. (2), respectively. Three DNA clones, P-061, atp6 and pSB8, were used as control probes in Southern hybridization and experiments. One of the genomic clones, P-061, is derived from nuclear DNA because F2 progeny segregated 1:2:1. The clones , atp6 and pSB8, are mitochondrial and chloroplast DNAs, respectively. These clones were obtained from rice by Kadowaki et al. (1990) and Hiratsuka et al. (1989). Southern hybridization was done according to Matsurra and Fujita (1994a).
Results. RFLP patterns of reciprocal hybrids. RFLP patterns of 11 paternal lines and 14 reciprocal hybrids are shown in Figure 1. Since these F1 hybrids produce biparental signals for P-061, these plants were considered hybrids. In contrast, all hybridization with B-174 and P-146 produced banding patterns like paternal lines in the reciprocal hybrids. Polymorphisms were observed between W-103 (lane No. 5: Cucumis sativus var. hardwickii) and other inbred lines when DNA was hybridized to atp6. Two reciprocal combinations which used W-103 as a parent showed the same signal as that of the paternal lines. Polymorphisms were not detected among these inbred lines when total DNAs were digested by BamHI and EcoRI and hybridized with pSB8.
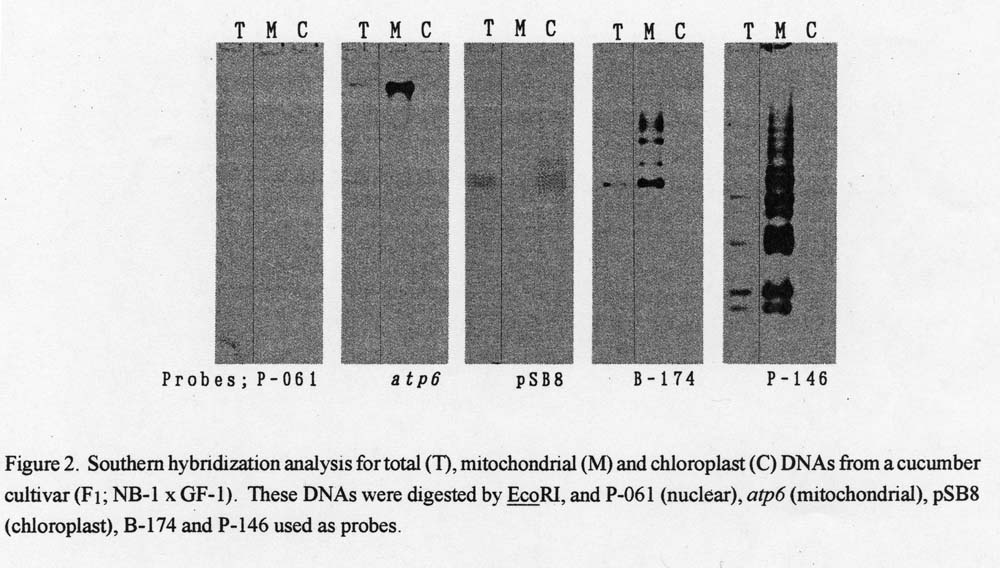
Origin of the two genomic clones. P-061, atp6 and pSB8 hybridized intensively with total mitochondrial and chloroplast DNAs, respectively (Fig. 2). Likewise, B-174 and P-146 hybridized intensively with mitochondrial DNA.
Discussion. These results suggest that the some parts of the mitochondrial DNA are inherited paternally in cucumber. Paternal inheritance of mitochondrial DNA has been reported in some plant species (8, 1). However, these species are distantly related to the Cucurbitaceae. In seed production of an F1 hybrid, cucumber breeders usually use gynoecious lines as the paternal parent. If some important agronomically characters are coded for in mitochondrial genes and polymorphisms were existed among the inbred lines, then attention must be given to the source of the paternal parent. It is important to determine whether these kind of polymorphisms exist in cultivated cucumber.
Figure 1. RFLP patterns resulting from Southern hybridization of RFLP clone P-061 to BamHI digested DNA, B-174 to Hind111-digested DNA, P-146 to EcoRI-digested DNA and atp6 to BamHI digested DNA which were isolated from 11 parents and 14 reciprocal hybrids.
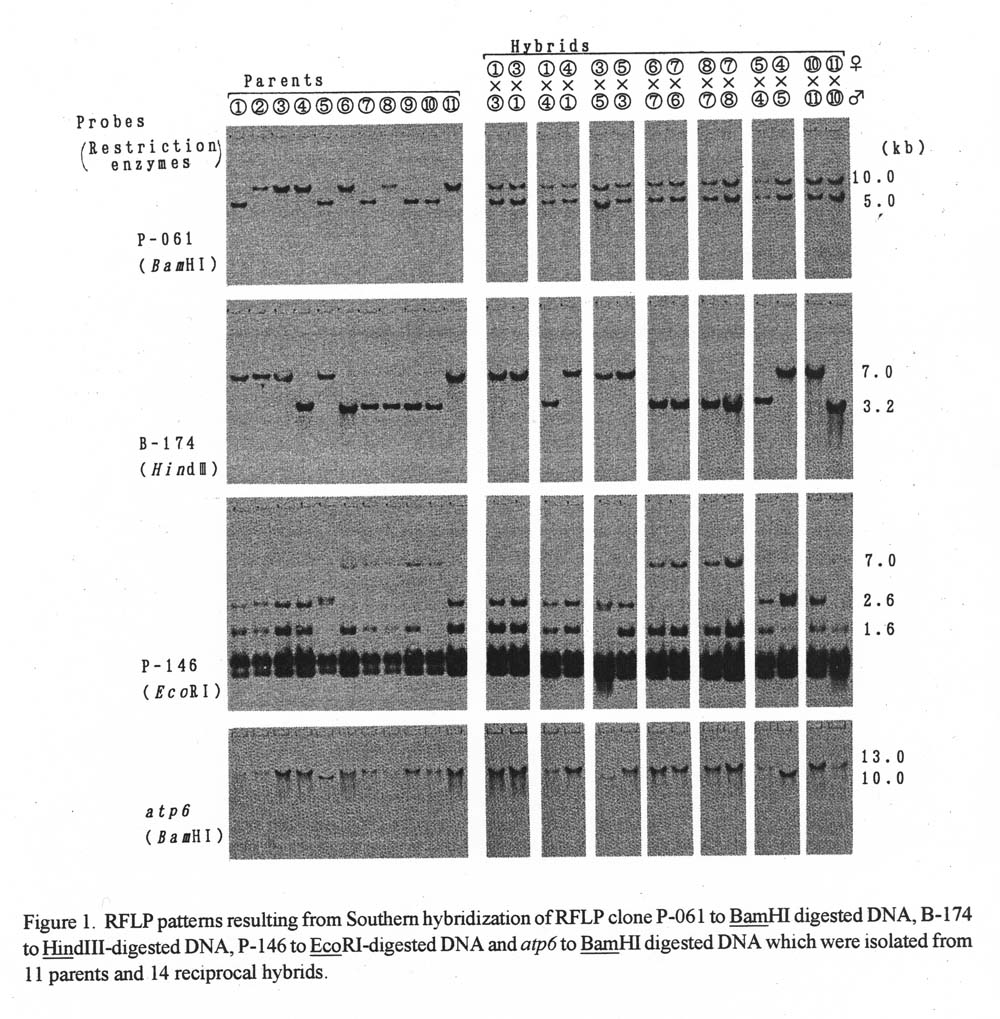
Figure 2. Southern hybridization analysis for total (T), mitochondrial (M) and chloroplast (C) DNAs from a cucumber cultivar (F1; NB-1 x GF-1). These DNAs were digested by EcoRI, and P-106 (nuclear), atp6 (mitochondrial), pSB8 (chloroplast), B-174 and P-146 used as probes.
Literature Cited
- Erickson, L. and R. Kemble. 1990. Paternal inheritance of mitochondria in rapeseed (Brassica napus). Mol. Gen. Genet. 222:135-139.
- Hirai, A., T. Ishibashi, A. Morikami, N. Iwatsuki, K. Shinozaki and M. Sugiura. 1985. Rice chloroplast DNA: a physical map and the location of the genes for the large subunit of ribulose 1.5-biphosphate carboxylase and the 32-kDa photosystem II reaction center protein. Theor. Appl. Genet. 70:117-122.
- Hiratsuka, J., H. Shimade, R. Whitter, T. Ishibashi, M. Sakamoto, M. Mori, C. Kondo,. Y. Honji, C-R. Sun, B.-Y. Meng, Y-Q. Li, A. Kanno, Y. Nishizaki, A. Hirai, K. Shinozaki, and M. Sugiura. 1989. The complete sequence of the rice (Oriza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol. Gen. Genet. 217:185-194.
- Kadowaki, K., T. Suzuki and S. Kazama. 1990. A chimeric gene containing the 5′ portion of atp6 is associated with cytoplasmic male-sterility of rice. Mol. Gen. Genet. 24:10-16.
- Matsuura, S. and Y. Fujita. 1994a. An approach for rapid checking of seed purity by RFLP analysis of nuclear DNA in F1 hybrid of cucumber (Cucumis sativus L.). J. Japan Soc. HOrt. Sci. 63:379-383.
- Matsuura, S. and Y. Fujita. 1994b. RFLP mapping of locus acr controlled sex expression in cucumber. In : Proceedings Cucurbitaceae 94: Evaluation and Enhancement of Cucurbit Germplasm, USDA/ARS, (in press).
- Murray, M. and W.F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 8:4321-4325.
- Neale, D.B., K.A. Marshall and R.R. Sederoff. 1989. Chloroplast and mitochondrial DNA are paternally inherited in Lequoia sempervirens D. Don Endl.. Proc. Natl. Acad. Sci. USA 86:9347-9349.
- Umbeck, P.K. and B.G. Gegenback. 1983. Reversion of male-sterile T-cytoplasm maize to male fertility in tissue culture. Crop Sci. 23:584-588.