Cucurbit Genetics Cooperative Report 21:25-28 (article 9) 1998
Y. Danin-Poleg, G. Tzuri, N. Regis and N. Katzir
Department of Vegetable Crops, Agricultural Research Organization, Northern Research Center, Newe Ya’ar, P.O.B. 9000 Haifa 31900, Israel
Introduction. Molecular marker techniques are beginning to have an impact on plant breeding and plant genetic resource management. A variety of methods have been employed to evaluate polymorphism in melon (Cucumis melo L.) including RFLP, RAPD, SSR and AFLP (1,3,4,5,6). Our studies on SSR have demonstrated that microsatellite repeats are abundant in the melon genome. We estimate that a GA/CT repeat occurs at approximately 200kb intervals (Danin-Polig et al., in preparation). This finding has led us to test the capacity of Inter-Simple Sequence Repeat (ISSR) markers (7) to discriminate melon cultivars. ISSR methods involve PCR amplification of DNA using a single primer composed of a microsatellite sequence anchored t the 3′ or 5′ end by 2-4 arbitrary, often degenerated, nucleotides. These primers target SSRs that are abundant throughout the genome and do not require prior knowledge of a DNA sequence. Amplification products can be separated on a polyacrylamide or agarose gel. Here we report the use of ISSR markers in distinguishing melon genotypes representing both wide and narrow genetic backgrounds.
Material and Methods. Eight Cucumis melo L. genotypes were tested PI 414723, ‘Dulce’, ‘Freeman’s Cucumber’, ‘Boy Yizre’el’, ‘Galia’, ‘Arava’ and ‘C8’ (Hegala) (the last three are F1 Galia types all bred at Newe Ya’ar). DNA was isolated from the bulked leaf or root tissue of 10 plants of each genotype using a mini preparation procedure described by Fulton et al. (2).
PCR reaction mixtures for separation on agarose gel (non-radiolabeled) contained: 30 ng of plant genomic DNA, 2 mM of MG2+ , 7.5pmole of primer(Kit #9, USA), 166 mM of each of the dNTPs, 1x Taq Buffer (Advanced Biotechnologies UK), ).5 unit of Taq DNA polymerase (Advanced Biotechnologies, UK), in a total volume of 25 ml.
The amplification program was as follows: 7 min at 94 C, 30 s at 94 C, 45 s at 45 C, and 2 min at 72 C for 35 cycles on a thermocycler (PTC-100 MJ Research Inc.).PCR products were separated by electrophoresis in a 1.2% agarose gel (Pechcomp LTD 9201) for a 4-5 hours and stained with ethidium bromide. The size marker used was PBR 322 Alw441/MVA1 (MBI Fermentas). PCR reaction mixtures for separation on sequencing gel (radiolabeled) contained 30 ng of plant genome DNA, lmM of Mg2+ , 7.5 pmole of primer, 277mM of dATP, dTTP, dGTP, 3.3mM of dCTP, 0.1 ml of 3000 CI/mmol [a-32P] dCTP or3000 Ci/mmol [a-33P] dCTP,1 x Taq Buffer (Advanced Biotechnologies, UL), 1 unit of Taq DNA polymerase (Advanced Biotechnologies, UK), in a total volume of 15 ml. The PCR conditions were the same as above but for 27 cycles. PCR products (3.0ml/lane) were separated on a DNA sequencing gel containing 6% polyacrylamide, 8M urea and 1x TBE, at 60 W constant power for 3.5-5.5 h. After drying, the gels were exposed to a Kodak XAR-5 film (Eastman Kodak). Forty-two ISSR primers were used to amplify DNA from the eight melon genotypes.
Results and Discussion. The majority of the primers (26 primers, 62%) detected polymorphism among the eight melon genotypes. Eight primers (19%) did not differ between melon genotypes, giving a monomorphic pattern, and eight (19%) primers failed to amplify a clear product.
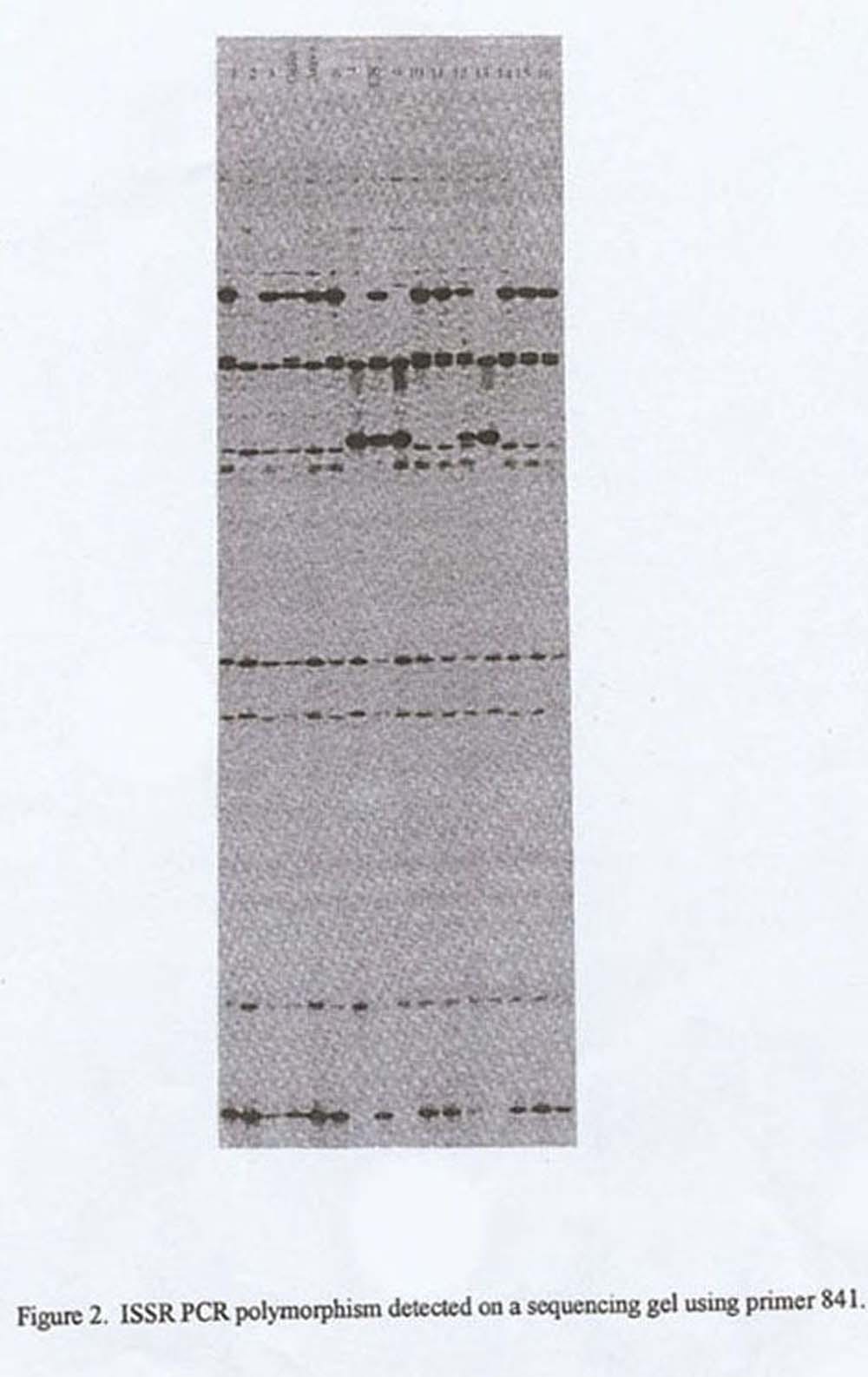
An average of 10 amplification products per primer for each genotype was separated on an agarose gel. Of these products 1 to 5 were clearly polymorphic. Amplification products of the primers that had yielded polymorphic patterns on an agarose gel were also tested on sequencing gels. The resolution of the sequencing gel was three times greater than that of the agarose gel. An average of 30 amplification products per primer for each genotype was detected on a sequencing gel with a minimum pf 10 polymorphic products. Figure 1 and 2 depict separation using an agarose and sequencing gel, respectively.
ISSR was applied to assess polymorphism between parental lines of a mapping population developed between PI 414723 and ‘Dulce’ (Cucumismelo var.acidulus Naud.) (3). Twenty one (50%) of the 42 ISSR primers that were tested distinguished between PI 414723 nd ‘Dulce’. For comparison, 16 (59%) of 27 melon SSR markers and 25 of 104 (24%) RAPD primers distinguished between the same two genotypes (Danin-Poleg, in preparation).
The efficiency of the ISSR method was further demonstrated by distinguishing among closely related genotypes. Twelve of the 42 primers (29%) detected polymorphism (yielding 21 polymorphic products) between two breeding lines within the Galia group, representing a narrow genetic background. Using one primer we can clearly distinguish among the cultivars Galia, Arava and C8 (Fig. 2). The majority of the bands amplified using ISSR primers segregate as dominant markers, yet some segregate as codominant markers when scored in a segregating population (for example see the band marked by an arrow in Fig. 2, where lanes 6 & 7 demonstrate the parental alleles while C8, the F1 between them clearly demonstrate the heterozygote.
The results here suggest that ISSR markers are highly polymorphic and informative in melon. They are especially valuable for diversity studies and legal protection where multi-loci markers are advantageous. In addition they may be efficiently used for map saturation.
Figure 1. ISSR PCR polymorphism detected on an agarose gel using primer 807. Lanes 5 and 6 are PI 414723 and ‘Dulce’, respectively.
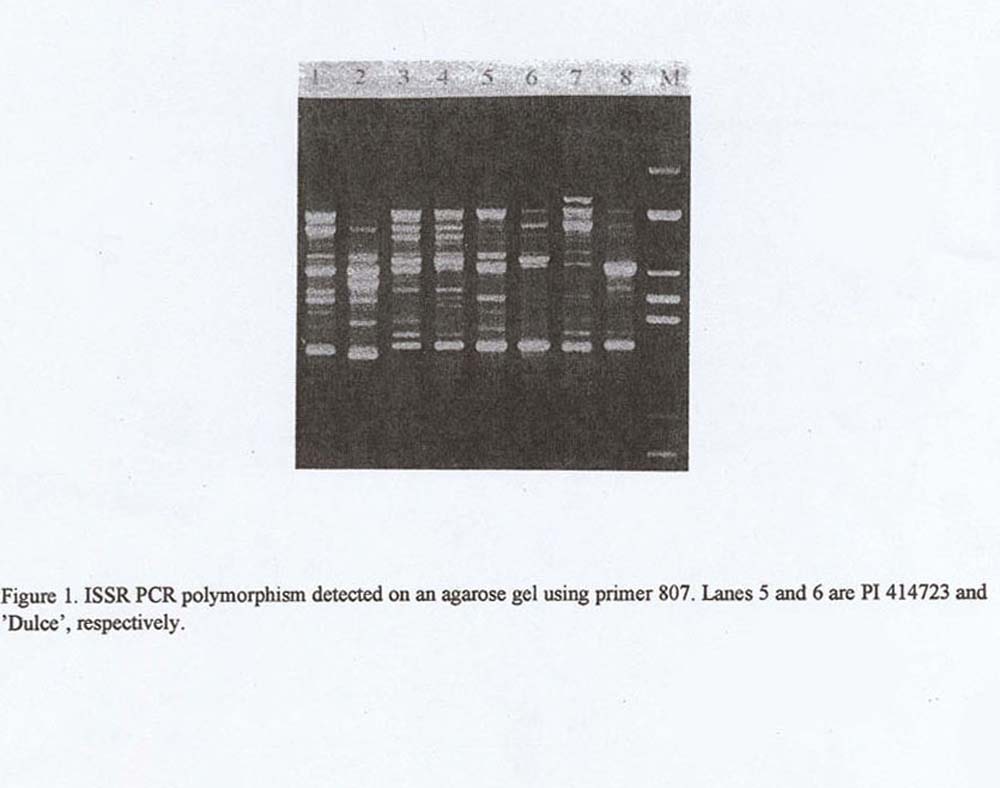
Figure 2. ISSR PCR polymorphism detected on a sequencing gel using primer 841.
Literature Cited
- Baudracco-Arnas, S. and M. Pitrat. 1996. A genetic map ofmelon (cucumis melo L.) with RFLP, RAPD, isozyme, disease resistance and morphological markers. Theor. Appl. Genet. 93: 57-64.
- Fulton, T.F., J. Chunwongse and S.D. Tankeley. 1995. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 13:207-209.
- Katzir N., Y. Danin-Poleg, G. Tzuri, Z.Karchi, U. Lavi and P.B. Cregan. 1996. Length polymorphism and homologies of microsatellites in several Cucurbitacceae species. Theor. Appl. Genet. 93:1282-1290.
- Neuhausen, L.S. 1992. Evaluation of restriction fragment length polymorphism in Cucumis melo. Theor. Appl. Genet. 83:379-384.
- Shattuck-Edens, D.M., R.N. Bell, S.L. Neuhausen and T. Helentjaris. 1990. DNA sequence variation within maize and melon: observations from polymerase chain reaction amplification and direct sequencing. Genetics 126:207-217.
- Wang, Y.H., C.E. Thomas and R.A. Dean. 1997. A genetic map of melon (Cucumis melo L.) based on amplified fragment length polymorphism (AFLP) markers. Theor. Appl. Genet. 95:791-798.
- Zietkiewicz, E., A. Rafalski and D. Labuda. 1994. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176-183.