Cucurbit Genetics Cooperative Report 7:27-30 (article 13) 1984
J.E. Staub and R.S. Kupper
Department of Horticulture, University of Wisconsin, Madison, WI 53706
T.C. Wehner
Department of Horticultural Science, North Carolina State University, Raleigh, NC 27695-7609
Evolutionary studies in the genus Cucumis have been conducted by Dane (2) and Esquinas-Alcazar (3) using isozymes as biochemical markers. In the study of Esquinas-Alcazar, electrophoretic variability was examined in 155 populations representing 21 species of Cucumis to determine intraspecific relationships within the species C. melo L. and interspecific relationships within the genus Cucumis. He used 6 enzymes [peroxidase (PRX), acid phosphatase (APR), glutamate oxalacetase transaminase (GOT), glucosephosphateisomerase (GPI), phosphoglucomutase (PGM), and 6- phosphoglucodehydrogenase (6-PDGH)] which were coded for a total of 16 enzyme-coding loci. The 21 species of Cucumis were classified, on the basis of genetic distances, into 4 groups. The anguria group included: C. africanus, C. anguria, C. ficifolius, C. myriocarpus, and C. zeyheri, while the sativus group included: C. sativus, C. hardwickii, and C. trigonus.
Two highly speculative hypotheses were proposed for the evolution of the sativus group. The hypotheses consisted of the following:
- The group could have been derived from an ancestral form of the metuliferus group through a primitive C. hardwickii following a reduction of chromosome number. This hypothesis is supported by the fact that the metuliferus group, which includes C. membranifolium, C. metuliferus and C. sagittatus is genetically close to the anguria and sativus groups. Moreover, Trivedi and Roy (8) concluded from cytogenetic studies that 12 might be the prime base chromosome number of Cucumis.
- The ancestral form of the metuliferus group and the sativus group could have had a common ancestor with 2n=14 chromosomes. This hypothesis implies that Cucumis species with a basic chromosome number x=12 evolved from those with 7 (1,4). Given that an unusual number of gene duplications have been observed between these groups (3), such duplication of genetic material could have taken place during the change in chromosome number.
In an earlier report (7), we documented the relative activity of 47 general metabolic enzymes and general protein in different tissues of selected botanical varieties of Cucumis [Cucumis sativus var. sativus L. and var. hardwickii (Royle) Kitamura]. In addition, we determined that enzyme polymorphisms existed in GPI, GR, isocitrate dehydrogenase (IDH), peptidase with phenyl-alanyl-proline (PEP- PAP), phosphogluconate dehydrogenase (PGD) and phosphoglucomutase (PGM). In this report we compare the electrophoretic phenotypes of several species of Cucumis for 16 enzymes [those mentioned above plus acid phosphatase (ACP), alkaline phosphatase (AKP), diaphorase (DIA), esterase (EST), fructose diphosphatase (FDP), glutamic pyruvic transaminase (GPT), leucine aminopeptidase (LAP), malate dehydrogenase (MDH), shikimic dehydrogenase (SKDH), and triose phosphate isomerase (TPI)] coding for 19 loci. The objective of this study was to compare zymograms of each of the loci to determine the magnitude of potential genetic difference between several species of the anguria and sativus groups as classified by Esquinas-Alcazar.
Cotyledonary extracts of 5 Cucumis species of African origin, 3 Cucumis sativus var. hardwickii and 6 var. sativus were examined by horizontal starch gel electrophoresis. Isozyme banding patterns of the enzymes were recorded and comparisons were made among zymograms (Table 1).
Table 1. Electrophoretic variation in 19 enzyme loci of several species in the genus Cucumis.
|
Species or botanical variety |
Cultivar name, inbred identification or PI no.z |
Sourcey |
Chromosome no. | Electrophoretic Phenotypes of Enzyme Locix | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acp | Akp | Dia-1 | Dia-2 | Est | Fdp | Gpi | Gpt | Gr-1 | Gr-2 | Idh | Lap | Mdh | Pep-pap | Pgd-1 | Pgd-2 | Pgm | Skdh | Tpi | ||||
|
C. africanus |
299570 |
S. Africa |
24 | 4 | 11 | 2 | 22 | 24 | 11 | 33 | 11 | — | 12 | 22 | 2 | 2 | 11 | 22 | 33 | 34 | 12 | 3 |
|
C. anguria |
147065 |
Brazil |
24 | 3 | 11 | 3 | — | 33 | 11 | 33 | 11 | — | 12 | 22 | 3 | 3 | 22 | 22 | 33 | 33 | 22 | 2 |
|
C. ficifolius |
196844 |
Ethiopia |
48 | 2 | 11 | 2 | — | — | — | 23 | 11 | — | 12 | 22 | 3 | 4 | — | 22 | 33 | 33 | 22 | 1 |
|
C. myriocarpus |
282447 |
S. Africa |
24 | 3 | 12 | 3 | 22 | 33 | 11 | 33 | 22 | 11 | 12 | 22 | 4 | 3 | 22 | 22 | 33 | 33 | 23 | 2 |
|
C. zeyheri |
282450 |
S. Africa |
24 | 3 | 12 | 3 | 22 | 33 | 11 | 33 | 22 | — | 12 | 22 | 4 | 3 | 22 | 22 | 33 | 33 | 23 | 2 |
|
C. sativus var. hardwickii |
183967 |
Umran, Kashia Hills, India |
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 11 | 11 | 1 | 1 | 22 | 11 | 11 | 11 | 22 | 1 |
|
215589 |
Dehra Dun, India
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 22 | 11 | 11 | 1 | 1 | 22 | 11 | 11 | 11 | 22 | 1 | |
|
462369
|
India
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 11 | 33 | 1 | 1 | 11 | 11 | 11 | 11 | 22 | 1 | |
|
C. sativus var. sativus
|
Marbel
|
Royal Sluis
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 11 | 33 | 1 | 1 | 11 | 11 | 11 | 11 | 22 | 1 |
|
Riesenschall
|
Royal Sluis
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 12 | 33 | 1 | 1 | 11 | 11 | 22 | 22 | 22 | 1 | |
|
GY 2*
|
NCSU
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 11 | 33 | 1 | 1 | 11 | 11 | 11 | 22 | 22 | 1 | |
|
1397*
|
USDA/ARS
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 11 | 11 | 33 | 1 | 1 | 11 | 11 | 22 | 22 | 22 | 1 | |
|
200815
|
Burma
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 11 | 33 | 12 | 11 | 33 | 1 | 1 | 12 | 11 | 11 | 11 | 22 | 1 | |
|
188807
|
Philippines
|
14 | 1 | 11 | 1 | 11 | 11 | 22 | 22 | 33 | 12 | 12 | 33 | 1 | 1 | 11 | 11 | 22 | 11 | 22 | 1 | |
zInbreds are identified by “*”.
yNCSU–North Carolina State University; USDA/ARS– United States Department of Agriculture/Agricultural Research Service.
xDiagramatic representation of phenotypes which have been given a single digit type number are provided in Figure 1.
The allelic nomenclature follows a modified form described by Richmond (5) such that loci coding for ACP, AKP, DIA, EST, FDP, GPI, GPT, GR, IDH, LAP, MDH, PEP-PAP, PGD, PGM, SKDH, and TPI are designated as Acp, Akp, Dia, Est, Fdp, Gpi, Gpt, Gr, Idh, Lap, Mdh, Pep-pap, Pgd, Pgm, Skdh, and Tpi, respectively. Hyphenated numerals refer to multiple loci, numbered from most cathodal to most anodal. Alleles at a particular locus are designated by numerals numbered from most cathodal to most anodal. As an example, the combination of homomeric protein products of the locus GR-1, which has at least 2 alleles (1 and 2), produce a heteromeric product in a heterozygous individual which is designated GR-1 (12). Where no immediate genetic interpretation of the zymograms could be made, individual zymogram patterns were given type numbers (Fig. 1).
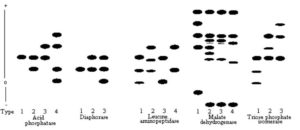
Figure 1. A diagramatic representation of electrophoretic variants observed in acid phosphatase, diaphorase, leucine aminopeptidase, malate dehydrogenase, and triose phosphate isomerase. Type numbers are given to identify each zymogram. The position of an isozyme does not reflect its relative mobility with regard to isozymes of other enzymes. Therefore, comparisons of relative isozyme mobilities should be made within and not between enzymes.
Electrophoretic phenotypes of all collections grouped as sativus were monomorphic for Acp, Dia, Est, Fdp, Gpt, Lap, Mdh, Pgd-1, Skah and Tpi, in which zymograms were single banded (Table 1). Phenotypic variation among botanical varieties was observed in Gpi, Gr-1, Gr-2, Idh, Pep-pap, Pgd-2 and Pgm. Those species grouped as anguria were monomorphic for Fdp, Gr-2, Idh, Pgd-1 and Pgd-2. If comparisons are made between groups, unique patterns (types) or alleles exist for Acp, Dia-1, Dia-2, Est,.Fdp, Gpi, Gpt, Idh, Lap, Mdh, Pgd-1, Pgd-2, Pqm and Tpi in the anguria group which do not appear in the sativus group.
These data indicate that, although species within the groups share some common alleles, enough difference exists between them to suggest that their genetic distance is certainly as great as Esquinas-Alcazar states. Moreover, comparisons among collections in the sativus group suggest that the genetic relationship between the two botanical varieties is probably much closer than gross morphological differences (6) would lead one to believe. These findings support the hypothesis of Dane (2) and Esquinas- Alcazar (3) regarding the conspecific nature of var. sativus and var. hardwickii. It would be interesting, with regard to the evolution of the genus Cucumis, to examine C. metuliferus and C. longipes in more detail in order to determine their potential relationship to C. sativus var. hardwickii and C. anguria.
Literature Cited
- Bhaduri, P.N. and P.C. Bose. 1947. Cytogenetical investigations in some common cucurbits with specific reference to fragmentation of chromosomes as a physical basis of speciation. J. Genet. 48:237-256.
- Dane, F.K. 1976. Evolutionary studies in the genus Cucumis. Ph.D. Thesis, Colorado State University, Fort Collins, Colorado.
- Esquinas-Alcazar, J.T. 1977. Alloenzyme variation and relationships in the genus Cucumis. Ph.D. Thesis, University of California, Davis, California.
- Kozuchov, Z.A. 1930. Karyological investigations of the genus Cucumis. (In Russian). Bul. Appl. Bot. Gen. and Plant Breeding 23:357-365.
- Richmond, R.C. 1972. Enzyme variability in the Drosophila williston group. 3. Amounts of variability in the superspecies D. paulistorum. Genetics 70:87-112.
- Schuman, D.A., J.E. Staub and B.E. Struckmeyer. 1982. Morphological comparisons between Cucumis sativus and Cucumis hardwickii plants. HortScience 17:108.
- Staub, J.E., R.S. Kupper and T.C. Wehner. 1983. Preliminary evaluation of isozyme polymorphisms in Cucumis. Cucurbit Gen. Coop. Rept. 6:32-34.
- Trivedi, R.N. and R.P. Roy. 1970. Cytological studies in Cucumis and Citrullus. Cytologia 36:561-569.