Cucurbit Genetics Cooperative Report 8:13-14 (Article 6) 1985
Ramachandran, C.
Agricultural University, Wageningen, 6708 PM, The Netherlands
The gene lists of different Cucumis species have been already compiled (3). However, the genes have not been assigned to any of the chromosomes, which are not distinguishable by normal staining procedures. Modern staining techniques like Giemsa C-banding has facilitated the identification of individual chromosomes in many plant and animal species (5). In this report, a standardised procedure for C-banding of chromosomes of Cucumis species by Giemsa staining is described. Essentially, the method suggested by Schwarzacher et al. (4) for orchids has been modified to suite the Cucumis chromosomes. The procedure involves the following steps:
- Germinate the seeds in a germinator at 28 to 30°C and collect the roots when they attain a 5 to 10 mm length. Meristematic roots can also be collected from the plants.
- Pre-treat the roots with 0.002 M hydroxyquinoline for 2.5 to 3 hr. at room temperature (20°C).
- Fix the roots in 1:3 acetic alcohol for 48 hr.
- Keep the roots in distilled water for 20 min. on the day of squashing.
- Transfer to 1 N HC1 for 5 min. at room temperature. – Macerate the roots in an enzyme solution containing 1% cellulase and 0.5% pectinase in 0.01 M citrate buffer (pH 4.0) at 38°C for 25 to 30 min.
- Soften the roots in 45% acetic acid for 25 to 30 min. at room temperature.
- Squash the meristematic tips in a drop of 45% acetic acid. Observe the chromosomes with a phase objective (Neofluar 40x) during slide preparation.
- Remove the coverslip after freeze drying in liquid nitrogen and air drying the slides for 2 days.
- Incubate the slides in 45% acetic acid at 60°C for 20 min.
- Denature the preparations in 6% Barium hydroxide solution for 6 to 7 min. at room temperature and wash in running tap water thoroughly for 1 hr.
- Incubate the slides in 2 x SSC (pH 6.8) at 60°C for 2 hr.
- Rinse the slides in 0.04 M phosphate buffer (pH 6.8) and stain in 4% Giemsa solution (in 0.04 phosphate buffer pH 6.8) for 10 to 15 min.
- Rinse the slides in phosphate buffer and air dry. Mount the preparation in euparol.
The procedure has yielded well differentiated banded complements in C. sativus var. hardwickii (Royle) Alef. (Fig. 1), C. sativus L. and C. melo L., which facilitated the identification of individual chromosomes in these species (1,2).
Acknowledgement
The financial support by the Dutch Organization for Advancement of Pure Research (ZWO) is gratefully acknowledged.
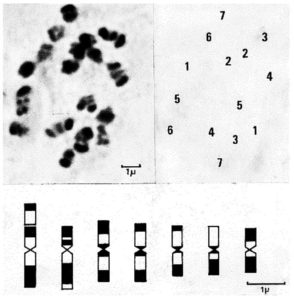
Fig. 1. Giemsa C-banded mitotic chromosomes and idiogram of C. sativus var. hardwickii (Royle) Alef.
Literature Cited
- Ramachandran, C and V. S. Seshadri. 1984. Cytogenetical analysis of the
genome of cucumber and muskmelon. Z. Planzenzuchtg. (in press). - Ramachandran, C., V.S. Seshadri and R.A. Pail 1983. Giemsa C-banding
in Cucumis. Proc. XV Intern. Congr. Genet. Contributory and Poster
PaperSession, New Delhi. (Abstr.) - Robinson, R.W. 1980. Known genes of Cucurbitaccae. Conference on the
Biology and Chemistry of Cucurbitaceae. Cornell Univ., Ithaca,
New York, U.S.A. - Schwarzacher, T., P. Ambros and D. Schweizer. 1980. Application of Giemsa
banding to orchid karyotype analysis. P1. Syst. Evol. 134:293-297. - Vosa, C.G. 1975. The use of Giemsa and other techniques in karyotype
analysis. Current Advances in Plant Science. 14:495-512.