Cucurbit Genetics Cooperative Report 9:18-23 (article 6) 1986
J.E. Staub
USDA, ARS, Department of Horticulture, University of Wisconsin, Madison, WI 53706
Electrophoretic, cucurbitacin and flavonoid variation; chromosome homology; and plant morphology have been studied to determine taxonomic relationships among cross-compatible species of Cucumis (1, 2, 4, 7, 8, 10, 11, 12, 15). These studies have furnished estimates of interspecific divergence which has led to increased prevision in the taxonomic classification of this genus. In a study in which the production, vigor, and fertility of F1 hybrids of 9 African species was used as a measure of intraspecific relationships, two species appeared to be closely related: C. prophetarum L. and C. anguria var. longipes A. Meeuse (personal communication, 10). Dane (2), was able to distinguish C. anguria var. anguria Meeuse and var. longipes supporting previous studies (3, 9) which proposed that these are varieties of a single species. Although, during an electrophoretic survey of 6 enzymes, Esquinas-Alcazar (4) failed to detect appreciable differences between these botanical varieties, we observed varietal differences while staining for peptidase with phenyl-alanyl-proline (PEP-PAP) and phosphoglucomutase (PGM) (unpublished data).
In an earlier report (13), we provided evidence that C. africanus Lindley F., C. anguria C. ficifolius A. Rich, C. myriocarpus Naud. and C. zeyheri Sond. were monomorphic for fructose diphosphatase, glutathione reductase, isocitrate dehydrogenase, and phosphogluconate dehydrogenase. Using a limited number of accessions, we gave a preliminary description of malate dehydrogenase (MDH) banding patterns in several AFrican species. It was thought desirable to acquire more information regarding the range of electrophoretic variation in African species for MDH. results from such a study could be used in designing future experiments which would test the genetic basis of the banding patterns observed and to examine evolutionary relationships. This communication describes the polymorphisms observed in 7 cucumis species for MDH and proposes a genetic model for testing this variation using cross-compatible species.
Cotyledons of 3 individuals of 7 C. Africanus, 11 C. anguria var. anguria, 4 C. anguria var. longipes, 4 C. dipsaceus Ehrenb. ex. Spach, 3 C. ficifolius, 2 C. metuliferus E. Mey ex,. Schrad., 3. C. myriocarpus, and 1 C. prophetarum L. collections were surveyed using horizontal starch gel electrophoresis. In order to standardize the relative mobilities of the observed electromorphs, extracts of the C. sativus L. inbred processing cucumber line Gy14A (X=7) were loaded on each gel, and band mobilities were recorded in relation to it. The least mobile (most cathodal) electromorph of Gy14A was designated 100 and all other bands were assigned a value based on their mobility relative to this electromorph.
A “Centroid” cluster analysis (5) was performed using the electromorphs observed to provide information on potential interspecific relationships not directly discernable from frequency data. Using this method, groups are depicted to lie in Euclidian space, and are replaced on formation by the co-ordinates of their centroid. The distance between groups is defined as the distance between the group centroids.
The frequencies of MDH electromorphs for each collection are given in Table 1. Each numeral indicates the number of times that a particular band was observed in 3 individuals of an accession. Although similarities among accessions are suggested by the presence of common electromorphs, their absence provides equivocal information since sample numbers were small. Bands which occur in C. anguria var. anguria are absent in var. longipes, possibly reflecting their varietal nature. Cucumis prophetarum possesses bands which are common to C. anguria var. longipes, but not to var. anguria. Several of the electromorphs recorded in C. anguria var. longipes were also observed in C. dipsaceus, C. ficifolius, and C. myriocarpus. Moreover, C. metuliferus, C. ficifolius, and C. myriocarpus shared some common electromorphs. Cluster analysis (Figure 1) may provide groupings which are meaningful if they parallel known biological facts. The classification of C. anguria var anguria accessions from Iran and those from other sources into two groupings is in agreement with a previous study (14). In contrast, the similarities observed in shikimic dehydrogenase, PEP-PAP, and PGM zymograms of C. metuliferus and C. myriocarpus were not recorded for MDH. Likewise, the lack of common electromorphs among C. myriocarpus and C. anguria would not have been predicted based on their ability to produce good to moderately self fertile hybrids (personal communication, A.P.M. den Nijs (10).
Table 1. The frequency of malate dehydrogenase electromorphs observed in 8 species of Cucumis.
Relative Electrophoretic Mobility of Maleate Dehydrogenase Electromorphsz |
||||||||||||||||||||
Species / Source |
95 |
97 |
98 |
99 |
100 |
104 |
106 |
107 |
109 |
110 |
113 |
115 |
118 |
125 |
127 |
131 |
132 |
133 |
135 |
138 |
| C. Africanus | ||||||||||||||||||||
| 273192 S. Africa | 3 | 3 | 3 | 3 | ||||||||||||||||
| 274036 “ | 2 | 1 | 1 | 2 | 2 | 2 | 2 | |||||||||||||
| 299569 “ | 3 | 3 | 3 | 3 | ||||||||||||||||
| 299570 Natal | 3 | 3 | 3 | 3 | ||||||||||||||||
| 299571 S. Africa | 3 | 2 | 3 | 2 | 2 | |||||||||||||||
| 299572 “ | 3 | 3 | 2 | 3 | 3 | |||||||||||||||
| 374151 USA | 1 | 1 | . | 3 | ||||||||||||||||
| C. anguria var. anguria | ||||||||||||||||||||
| 147605 Brazil | 3 | 2 | 1 | 1 | ||||||||||||||||
| 196477 “ | 3 | 2 | 1 | 2 | 1 | 2 | 2 | |||||||||||||
| 233646 Ethiopia | 3 | 3 | 1 | 3 | ||||||||||||||||
| 386029 Iran | 3 | 2 | ||||||||||||||||||
| 386031 “ | 3 | |||||||||||||||||||
| 386034 “ | 3 | |||||||||||||||||||
| 390449 Ecuador | 3 | 3 | 2 | 1 | ||||||||||||||||
| 438570 Guatemala | 3 | 3 | 3 | 1 | 3 | |||||||||||||||
| 438678 Mexico | 3 | 3 | 3 | 3 | 3 | |||||||||||||||
| 438679 “ | 3 | 3 | 2 | 1 | 3 | |||||||||||||||
| 442176 “ | 3 | 2 | 2 | 3 | ||||||||||||||||
| C. anguria var. longipes | 0 | |||||||||||||||||||
| 249894 S. Rhodesia | 3 | 2 | 1 | 1 | ||||||||||||||||
| 249895 “ | 3 | |||||||||||||||||||
| 249896 “ | 1 | 2 | ||||||||||||||||||
| 249897 “ | 3 | 3 | 1 | 3 | ||||||||||||||||
| C. dipsaceus | ||||||||||||||||||||
| 193498 Ethiopia | 3 | 1 | 2 | 1 | ||||||||||||||||
| 236468 “ | 3 | 1 | ||||||||||||||||||
| 390450 Ecuador | 3 | 2 | 1 | 2 | ||||||||||||||||
| 441993 Netherlands | 3 | 2 | 1 | 1 | 1 | 2 | ||||||||||||||
| C. myriocarpus | ||||||||||||||||||||
| 203977 S. Africa | 3 | 2 | 2 | 3 | 3 | |||||||||||||||
| 282447 “ | 3 | 2 | 3 | |||||||||||||||||
| 282449 “ | 3 | 1 | 1 | 2 | ||||||||||||||||
| C. sativus | 1 | |||||||||||||||||||
| Gy14A USA | 3 | 3 | ||||||||||||||||||
| C. ficifolius | ||||||||||||||||||||
| 196844 Ethiopia | 3 | 2 | 3 | |||||||||||||||||
| 273648 “ | 3 | 2 | ||||||||||||||||||
| 280231 “ | 3 | 3 | ||||||||||||||||||
| C. metuliferus | ||||||||||||||||||||
| 202681 S. Africa | 3 | 1 | 3 | 2 | 1 | |||||||||||||||
| 292190 Transvaal | 3 | 3 | 3 | |||||||||||||||||
| C. prophetarum | ||||||||||||||||||||
| 193967 Ethiopia | 1 | 2 | 1 | 1 | ||||||||||||||||
| z Electromorph with the least mobility in C. sativus (Gy14A) was designated as 100. All other electromorphs classified relative to 100. | ||||||||||||||||||||
| y Frequency of electromorph observed with an accession. | ||||||||||||||||||||
| x R.L. Lower University of Wisconsin. | ||||||||||||||||||||
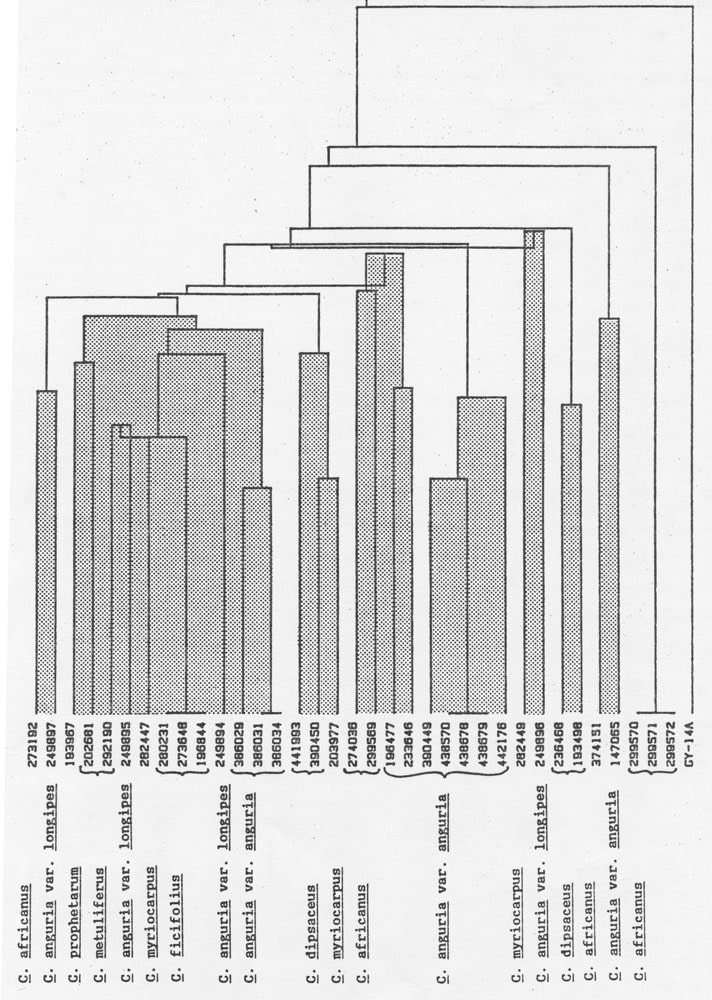
Figure 1. Cluster analysis of 8 Cucumis species based on electrophoretic variation of malate dehydrogenase.
It would be desirable to determine the genetic basis of MDH variation and thereby use allelic frequencies (allozymes) to identify relationships among species. The fertility relationships among C. myriocarpus, C. prophetarum, and C. anguria var. anguria and var. longipes offer an avenue in which a genetic hypothesis of this dimeric enzyme (6) could be tested. Since these species have several electromorphs in common (Figure 2), isozyme patterns of individuals of each species could be identified, vegetative cuttings made and testing of a proposed hypothesis. From the data provided by the present study, it would be reasonable to hypothesize a simple one locus model with 5 alleles. This hypothesis is based on the fact that all electromorphs became visible at approximately the same time and the presence of electromorphs located approximately midway between other electromorphs. Studies are being designed which test this hypothesis.
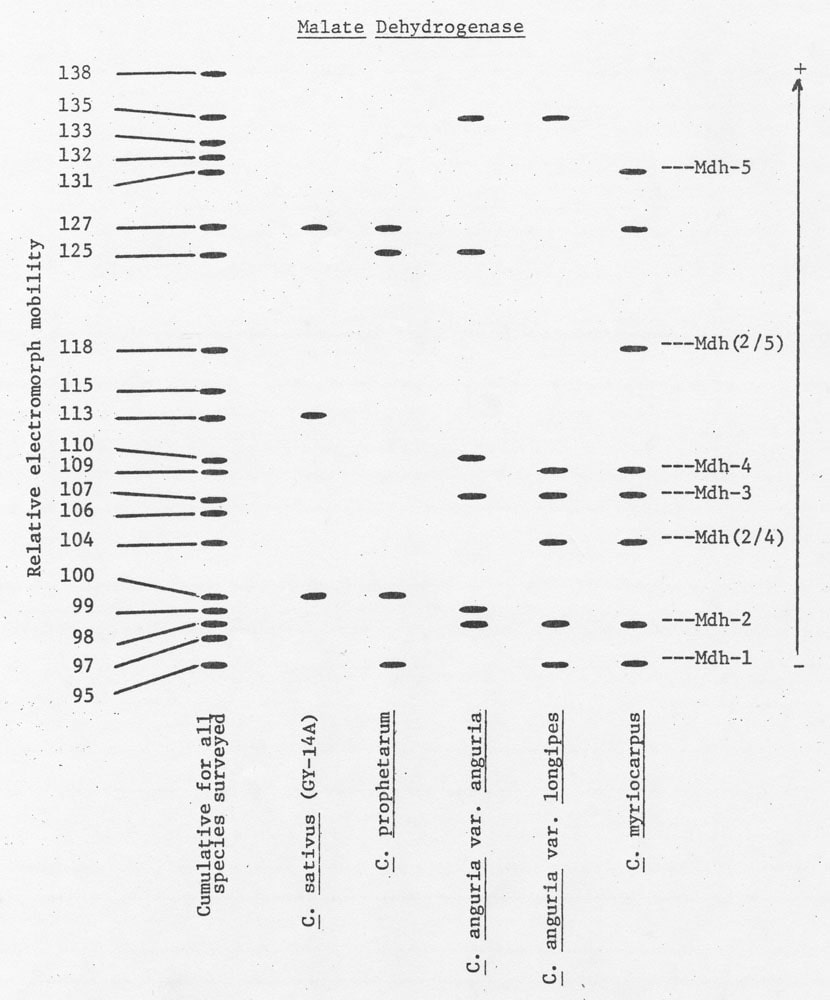
Figure 2. A diagramatic representation of electrophoretic variants of malate dehydrogenase observed in a preliminary survey. The possible position of homo- and heterodimers of a hypothetical model for the explanation of observed isozyme variation is also presented.
Literature Cited
- Brown, G.B., J.R. Deakin, and M.B. Wood. 1969. Identification of Cucumis species by paper chromatography of flavonoids. J. Amer. Soc. Hort. Sci. 94:231-234.
- Dane, F. 1983. Cucurbits. In: Isozymes in plant genetics and breeding Part B, Pg. 369-390. Edited by S.D. Tanksley and T.J. Orton. Amsterdam: Elsevier.
- Deakin, J.R., G.W. Bohn, and T.W. Whitaker. 1971. Interspecific hybridization in Cucumis. Econ. Bot. 25:195-211.
- Esquinas-Alcazar, J.T. 1977. Alloenzyme variation and relationships in the genus Cucumis. Ph.D. Dissertation, University of California, Davis, California, 170 pp.
- Everitt, B. 1974. Cluster analysis. Halsted Press, Division of John Wiley & Sons, Inc. New York, pp 110.
- Harris, H. 1974. Multiple allelism and isozyme diversity in human populations. In: Isozymes. IV: Genetics and Evolution. Ed. C.L. Markert. Academic Press. New York, pp. 964.
- Kho, Y.O., A.P.M. den Nijs, and J. Franken. 1980. Interspecific hybridization in Cucumis L. II. The crossability of species, an investigation of in vivo pollen tube growth Euphytica 29:661-672.
- Kozuchov, Z.A. 1930. Karyological investigations of the genus Cucumis. Bull. Appl. Bot. Gen. Plant Breed. 23:357-365.
- Meeuse, A.D.J 1958. The possible origin of Cucumis anguria L. Blumea Suppl. IV. 196-204.
- Nijs, A.P.M. den, and K.D.L. Visser. 1985. Relationships between African species of the genus Cucumis L. estimated by the production, vigour and fertility of F1 hybrids. Euphytica (in press).
- Rehm, S. 1960. Die bitterstoffee der cucurbitacean. Erg. Biol. 22:108-136.
- Singh, A.K. and K.S. Yadava. 1984. An analysis of interspecific hybrids and phylogenetic implications in Cucumis (Cucurbitaceae). Pl. Syst, Evol. 147:237-252.
- Staub, J.E. and R.S. Kupper, 1984. Electrophoretic comparison of six species of Cucumis. Cucurbit Genetics Cooperative Rpt. 7:27-30.
- Staub, J.E. and L. Fredrick. 1985. Electrophoretic variation among wild species in the genus Cucumis. Cucurbit Genetics Cooperative Rpt. 8:22-25.
- Whitaker, T.W. 1933. Cytological and phylogenetic studies in the Cucurbitaceae. Bot. Gaz. 94:780-790.