Cucurbit Genetics Cooperative Report 10:4-6 (article 3) 1987
C. Ramachandran
College of Horticulture, Kerala Agricultural University, Vellanikkara 680651, India
R.K.J. Narayan
Department of Agricultural Botany, University College of Wales, Aberystwyth, U.K.
The genus Cucumis comprises about thirty species of Asian and African origin. The diploid species are dibasic with haploid chromosome numbers, n=7 or n=12. About 17 percent of Cucumis species are polyploids and their chromosome numbers vary from 4x=48 to 6x=72. The 2C DNA amounts among diploid species range narrowly between 1.37 pg and 2.48 pg, and polyploids between 2.48 pg and 3.38 pg. The quantitative DNA changes associated with speciation in Cucumis has affected all chromosomes within each chromosome complement (5).
The nuclear DNA composition of four Cucumis species viz., C. melo, C. metuliferus, C. anguria and C. sativus were studied by buoyant density gradient analysis and thermal denaturation analysis. The method for DNA extraction was similar to that described by Bendich and Anderson (1). Analytical CsCl density gradient analysis was done in a Centriscan – 75 ultracentrifuge. Purified DNA (1.5 mg from each Cucumis species) was centrifuged to equilibrium in neutral CsCl (density 1.710 g/ml). Micrococcus lysodeikticus DNA of known buoyant density was included as a marker. For thermal denaturation analysis, DNA samples were melted in 0.1 x SSC (SSC = 0.15M NaCl + 0.015M trisodium citrate, pH 7.0) in a fully automated SP 1800 Spectrophotometer equipped with an electronically heated cell block. The increase in absorbance (hyperchromicity) was recorded automatically in a digital printout for every 0.25°C increase in temperature. The absorbance measurements after correction for solvent expansion (4) were plotted as a ratio, At /A25 (absorbance at temperature ‘t’ divided by initial absorbance at 25°C) against temperature.
The buoyant density profiles for all species were asymmetric (Figure 1). The asymmetry suggests the presence of satellite DNA sequences with different average buoyant density. Satellite DNA sequences have been identified and characterized previously in the genomes of C. sativus and C. melo (1,2,3). The buoyant densities for the total main band DNA ranged from 1.6909 g/ml for C. anguria to 1.6920 g/ml for C. sativus, C. melo and C. metuliferus have intermediate values (Table 1). The melting profiles when compared under strictly identical ionic conditions gave us information about the base ratio of DNA as well as about heterogeneity, if any, in the dispersion of base pairs. Tm (the temperature corresponding to half the final increase in relative absorbance), base ratio (G+C content) and base compositional heterogeneity estimated from the melting profile are also given in Table 1. Tm values ranged between 69.5°C and 70.25°C. Guanine + Cytosine content (G+C) of DNA estimated from Tm was similar to the values derived from buoyant density (7) and ranged from 38.06 to 39.89%. C. sativus DNA has an entirely different estimate of base compositional heterogeneity (14.67%) compared with the other three Cucumis species. Detailed results of the isolation, characterization and in situ hybridization of satellite DNA sequences from Cucumis species will be published elsewhere (6).
Table 1. Nuclear DNA composition of Cucumis species
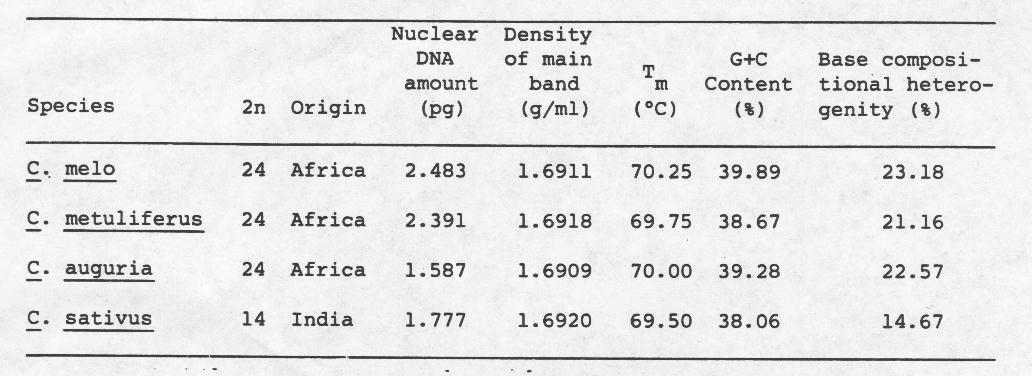
Figure 1. Buoyant density profiles for nuclear DNA of Cucumis species with marker DNA.
Literature Cited
- Bendich, A.J. and R.S. Anderson. 1974. Novel properties of satellite DNA from muskmelon. Proc. Natl. Acad. Sci. USA. 71:1511-1515.
- Hemleben, V., B. Leweken, A. Roth and J. Stadler. 1982. organization of highly repetitive DNA of two Cucurbitaceae species (Cucumis melo and Cucumis sativus). Nucleic Acid Research 10:631-644.
- Ingle, J., J.N. Timmis and J. Sinclair. 1975. The relationship between satellite DNA, RNA, gene redundancy and genome size in plants. Plant Physiol. 55:496-501.
- Mandel, M. and J. Marmur. 1968. Use of ultraviolet absorbance-temperature profile for determining the guanine plus cytosine content of DNA. Methods Enzymol. 12(B):195-206.
- Ramachandran, C. and R.K.J. Narayan. 1985. Chromosomal DNA variation in Cucumis. Theor. Appl. Genet. 69:497-502.
- Ramachandran, C. and R.K.J. Narayan. 1986. Satellite DNA specific to knob heterochromatin in Cucumis metuliferus. Mol. Gen. Genet. (In press).
- Schilkraut, C.L., J. Marmur and P. Doty. 1962. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J. Mol. Biol. 4:430-443.