Cucurbit Genetics Cooperative Report 9:27-32 (article 8) 1986
Staub, J.E., R. Kane and R. S. Kupper
U.S. Department of Agriculture, Agricultural Research Service and Department of Horticulture, University of Wisconsin, Madison, WI 53706
Fruit yield in cucumber (cucumis sativus L.) is suppressed due to the physiological nature of its fruiting habit. Fruit developing from the firs pollinated flower inhibits the development of subsequent fruits. It is unclear whether fruit set inhibition is due to a substance translocated from the fruit, a substrate limited source-sink relationship, or some other physiological mechanism (3, 5, 7, 8, 14).
The incorporation of quantitatively inherited characters into commercially adapted cultivars using exotic germplasm can be an effective way to obtain greater genetic variability and response to selection (1, 4). Cucumis sativus var. hardwickii (R.) Alef., a multiple fruiting phenotype, has been suggested as a potential source for increasing the genetic variability for yield in cucumber since it lacks fruit set inhibition (6,11).
One thrust of the USDA cucumber breeding program involves the utilization of hardwickii germplasm in cucumber improvement. Potentially useful multiple fruiting populations derived from initial hardwickii x sativus matins are being developed which possess multiple disease resistance along with high levels of gynoecy and the non-b9tter character (12m13). Data indicate that there are morphological and anatomical differences among several of the hardwickii plant introductions used in development of these populations (10). We have used two hardwickii collections in our breeding program, PI 215589 and LJ 904030. It would be useful to determine if these and other hardwickii collections could be grouped, thus providing more efficient use of this germplasm. Therefore, an experiment was designed to characterize differences among 7 hardwickii accessions currently available for use in plant improvement.
Three diverse sativus gynoecious inbred lines (WI 1379, WI 1909. and GY-14) were selected for crossing with 7 lines of hardwickii (LJ 90430, LJ 91176, PI 183967, PI 215589, PI 273648, PI 462369, PI 486336). A Design II mating scheme (2) was initiated by crossing each sativus line with each hardwickii line to produce 21 F1 families (3×7). This mating scheme was used to negate the potential photoperiodic responses associated with hardwickii (13) and to provide information on the combining ability of the hardwickii collections used. The F1 progenies were planted in field nurseries at the University of Wisconsin experimental stations at Hancock and Arlington, Wisconsin. The soil type at Hancock is Planefield loamy sand (Typic Udipsamment; sandy, mixed, mesic) while the Arlington soil type is Plano silt loam (Typic Argiudoll; fine-silty, mixed, mesic). Three and 6 replications were arranged in a randomized complete block design at Arlington and Hancock, respectively. In each block, 9 individuals of each cross were spaced 1.52 m apart within a row, and parallel rows were designated as plot borders. Data were collected from the 7 innermost plants of a row. Border rows of similar lineage were also planted on the outside of each block. Supplemental irrigation was used along with standard cultural practices.
Data from each plant within a plot were collected on the number of days to anthesis, number of female nodes, fruit number, length and diameter, number of lateral branches, and plant dry weight. Fruit length/diameter ratios (D\L/D) were also calculated. For each parameter, measurements of the 7 plants within a plot were averaged and these means were the experimental units used for analysis. Cumulative means (over replications) for each series of F1 progeny are presented in Tables 1 and 2.
An ordination of variables by principal component analysis (PCA) was performed in order to aid in the interpretation of the multivariate data (9). The ordination obtained by PCA allows for the groupings of hardwickii collections into subpopulations based on their relative differences. Although in PCA one woRks from the data towards a hypothetical model (grouping into subpopulations), PCA should be considered an initial step in which complex data sets are simplified to make them more amenable to interpretation. Principal component analysis is an analytical procedure for transforming one set of variates into another set of component varieties which are linear, orthogonal functions (eigenvectors) of the original variates. The total variation is equal to the total variation of the original variates such that the variance associated with each component decreases in order (i.e., the first variate will account for the largest possible proportion of the total variation, the second will account for the largest portion of the remainder and so forth). An assumption basic to PCA is that the observed variation is caused by the effects that the underlying (causal) factors have on each of the original variates. The consistency of these underlying factors on character expression and therefore on ordination (the ordering of units within a multidimensional space) can be examined by comparison of data from different locations.
The intravarietal relationships among hardwickii plant introductions was examined by casting the eigenvectors and associated components derived from F1 data into a multidimensional hyperspace (9; Figure 1). Location data are presented separately such that the X-axis corresponds to the first eigenvector and the Y-axis to the second eigenvector for each location. The distance among these points is proportional to the degree of dissimilarity in terms of the set of variates (parameters) used. If discrete subpopulations with some degree of biological integrity can be defined, then hardwickii accessions can be classified.
Analysis indicates that PI 183967 and LJ 90430 could be grouped into a distinct subpopulation, while PI 273648 and LJ 91176 could form another. Although PCA of Arlington data indicate that PI 215589 and PI 486336 show similarities, analysis of Hancock data provides no such representation. The accession, PI 486336, appears to be most similar to PI 273648 and therefore bears some resemblance to LJ 91176. Principle component analysis of both locations revealed that PI 462369 could be classified as a unique subpopulation.
The grouping of PI 183967 and LJ 90430 is consistent with the fact that LJ 90430 is a derived line from the intermating PI 183967 (personal communication, J.D. McCreight). The PI 462369 was obtained from the V.I. Vavilov Institute of Plant Breeding, USSR and is itself closer to sativus than any other accession examined with regard to fruit characteristics. These fruit characteristics appear to be transferable (apparent in F1 progeny) and undoubtedly contribute to the ordination of this accession. The PI 215589 was collected near Dehra Dun, India (H.S. Gentry, 1954) while PI 486336 was acquired on Mount Abu, India (personal communicat9ion, B. Dutt). These collection sites are, ecologically speaking, very dissimilar. It might be reasonable to expect that the performance of F1 progeny derived from these accessions would be dissimilar in different locations.
These data indicate that there are differences among F1 progeny derived from hardwickii x sativus matings. The 8 parameters monitored at 2 locations allowed for the discrimination of 7 hardwickii parental accessions into 3 distinct subpopulations by PCA. Although any hypothesis obtained from such an analysis must be considered subjective until confirmed by additional studies, it is likely from these and other data (12) that the hardwickii accessions used in this study are not similar and that their utilization in a plant improvement program will be dictated by project objectives.
Table 1. Means by cross of morphological traits of F1 hybrids of Cucumis sativus L. and C. sativus var. hardwickii (R.) Alef. at Hancock, WI.
Fruit |
|||||||||
Cross |
Fruit no. |
Lateral branch no. |
Plant dry wt. (R) |
Length (cm) |
Diameter (cm) |
Length/diameter ratio |
No. of female nodesz |
No. of days to anthesis |
|
| GY14 x | LJ 90430 | 62.84 | 11.86 | 253.63 | 8.97 | 5.30 | 1.69 | 0.260 | 40.42 |
| LJ 91176 | 55.35 | 11.17 | 292.61 | 9.26 | 5.79 | 1.60 | 0.318 | 41.64 | |
| PI 183967 | 73.17 | 11.90 | 285.62 | 9.18 | 5.32 | 1.72 | 0.284 | 40.49 | |
| PI 215589 | 34.79 | 10.67 | 206.79 | 9.60 | 5.85 | 1.64 | 0.366 | 42.61 | |
| PI 273648 | 44.82 | 11.31 | 280.79 | 10.02 | 6.14 | 1.63 | 0.259 | 41.04 | |
| PI 462369 | 25.92 | 9.21 | 201.46 | 12.61 | 6.27 | 2.01 | 0.264 | 38.84 | |
| PI 486336 | 57.59 | 11.68 | 282.87 | 9.58 | 5.97 | 1.60 | 0.330 | 39.10 | |
| WI 1379 x | IJ 90430 | 62.84 | 10.73 | 248.49 | 9.04 | 5.44 | 1.66 | 0.281 | 39.57 |
| IJ 91176 | 52.48 | 103.25 | 257.07 | 9.28 | 5.71 | 1.63 | 0.275 | 40.21 | |
| PI 183967 | 62.93 | 11.48 | 239.37 | 9.01 | 5.34 | 1.69 | 0.274 | 39.86 | |
| PI 215589 | 44.02 | 10.74 | 229.95 | 9.46 | 5.60 | 1.69 | 0.353 | 37.22 | |
| PI 273648 | 44.90 | 10.41 | 240.40 | 9.70 | 5.93 | 1.63 | 0.265 | 39.69 | |
| PI 462369 | 23.09 | 8.46 | 176.52 | 12.78 | 6.32 | 2.02 | 0.307 | 38.12 | |
| PI 486336 | 52.24 | 10.55 | 246.85 | 9.40 | 5.84 | 1.61 | 0.348 | 37.92 | |
| WI 1909 X | LJ 90430 | 63.64 | 11.09 | 233.28 | 10.26 | 5.20 | 1.97 | 0.336 | 40.33 |
| LJ 91176 | 52.71 | 10.40 | 259.52 | 10.62 | 5.48 | 1.94 | 0.316 | 39.83 | |
| PI 183967 | 56.95 | 10.85 | 243.06 | 10.56 | 5.16 | 2.05 | 0.338 | 40.52 | |
| PI 215589 | 35.80 | 9.88 | 185.49 | 11.02 | 5.68 | 1.94 | 0.375 | 41.43 | |
| PI 273648 | 43.36 | 10.55 | 244.42 | 10.90 | 5.80 | 1.88 | 0.325 | 40.02 | |
| PI 462369 | 24.25 | 8.37 | 192.18 | 14.18 | 6.10 | 2.32 | 0.292 | 39.19 | |
| PI 486336 | 48.53 | 10.60 | 227.28 | 10.21 | 5.68 | 1.80 | 0.338 | 39.55 | |
| Grand Mean | 48.64 | 10.58 | 239.42 | 10.27 | 5.71 | 1.80 | 0.310 | 37.87 | |
z Arcsine squareroot transformation of the data.
Table 2. Means by cross of morphological traits of F1 hybrids of Cucumis sativus L. and C. sativus var. hardwickii (R.) Alef,. at Arlington, WI.
Fruit |
|||||||||
Cross |
Fruit no. |
Lateral branch no. |
Plant dry wt. (g) |
Length (cm) |
Diameter (cm) |
Length/diameter ratio |
No. of female nodesz |
No. of days to anthesis |
|
| GY 14 x | IJ 90430 | 114.83 | 11.62 | 436.97 | 8.80 | 5.13 | 1.71 | 0.256 | 49.70 |
| IJ 91176 | 64.76 | 8.71 | 274.48 | 8.77 | 5.71 | 1.53 | 0.295 | 51.57 | |
| PI 183967 | 78.09 | 11.14 | 363.29 | 8.79 | 5.04 | 1.74 | 0.223 | 53.29 | |
| PI 215589 | 46.61 | 9.05 | 186.62 | 9.05 | 5.64 | 1.60 | 0.355 | 48.39 | |
| PI 273648 | 53.66 | 10.02 | 287.54 | 9.99 | 6.20 | 1.61 | 0.244 | 50.12 | |
| PI 462369 | 22.03 | 7.31 | 105.80 | 12.27 | 6.30 | 1.95 | 0.253 | 49.71 | |
| PI 486336 | 46.63 | 9.35 | 142.58 | 8.75 | 5.62 | 1.56 | 0.319 | 50.22 | |
| WI 1379 X | LJ 90430 | 81.52 | 10.95 | 260.59 | 8.39 | 5.11 | 1.64 | 0.286 | 48.95 |
| LJ 91176 | 48.99 | 7.95 | 215.38 | 8.87 | 5.64 | 1.57 | 0.262 | 51.09 | |
| PI 183967 | 81.12 | 9.56 | 254.05 | 8.69 | 5.07 | 1.71 | 0.294 | 48.43 | |
| PI 215589 | 42.28 | 8.44 | 148.60 | 9.19 | 5.66 | 1.62 | 0.294 | 47.81 | |
| PI 273648 | 46.33 | 8.38 | 195.47 | 9.28 | 5.85 | 1.58 | 0.371 | 51.05 | |
| PI 462369 | 17.67 | 6.11 | 96.20 | 12.20 | 6.28 | 1.94 | 0.300 | 47.33 | |
| PI 486336 | 54.28 | 8.70 | 186.47 | 9.03 | 5.77 | 1.56 | 0.308 | 47.29 | |
| WI 1909 | LJ 90430 | 73.77 | 10.54 | 307.43 | 9.74 | 5.04 | 1.93 | 0.330 | 49.96 |
| LJ 91176 | 41.86 | 8.54 | 209.18 | 10.24 | 5.43 | 1.89 | 0.274 | 54.19 | |
| PI 183967 | 79.51 | 11.27 | 355.69 | 10.12 | 4.93 | 2.05 | 0.341 | 50.84 | |
| PI 215589 | 37.78 | 9.04 | 126.53 | 9.97 | 5.53 | 1.80 | 0.376 | 51.83 | |
| PI 273648 | 52.12 | 10.86 | 264.53 | 10.75 | 5.81 | 1.85 | 0.349 | 52.00 | |
| PI 462369 | 19.79 | 6.97 | 144.76 | 13.40 | 5.87 | 2.28 | 0.327 | 50.62 | |
| PI 486336 | 42.64 | 8.66 | 159.20 | 10.03 | 5.49 | 1.82 | 0.361 | 53.16 | |
| Grand Mean | 54.57 | 9.20 | 224.81 | 9.82 | 5.58 | 1.76 | 0.308 | 50.36 | |
zArcsine squareroot transformation of the data.
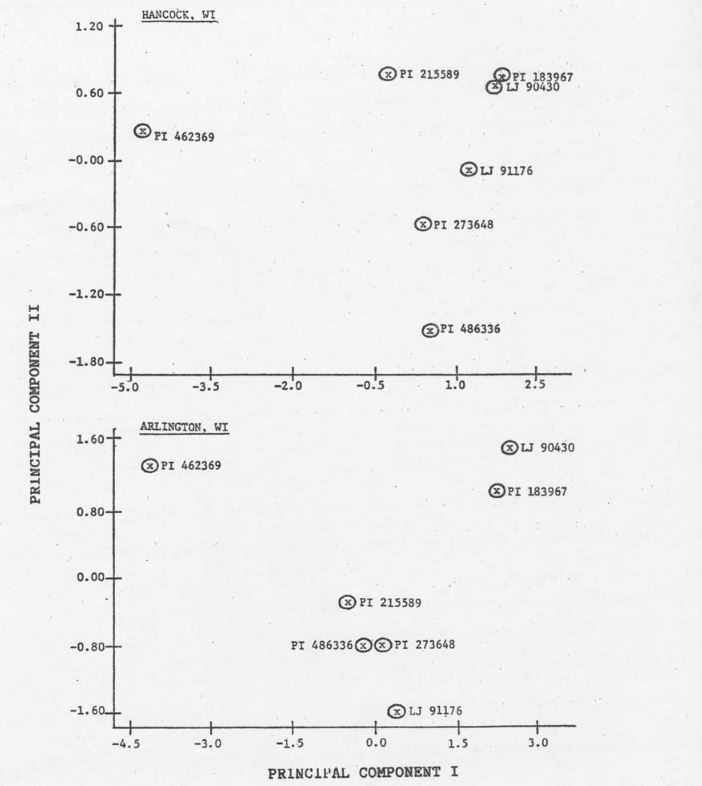
Figure 1. Plot of individual C. sativus var. hardwickii (R) Alef. accessions based on scores of F1 progeny (C. sativus var. sativus x war. hardwickii) on principal component axes I and II at two locations. Each point represents the measurement of 8 parameters in each of 6 replications per location.
Literature Cited
- Bliss, F.A. 1981. Utilization of vegetable germplasm. HortSci. 16:129-132.
- Comstock, R.E. and H.F. Robinson. 1948. The components of genetic variance in populations of biparental progenies and their use in estimating the average degree of dominance. Biometrics 4:254-266.
- Deena, D.W. 1973. The effects of genetic parthenocarpy and gynoecious flowering habit on fruit production and growth in cucumbe, Cusumis sativus L. J. Amer, Soc. Hort. Sci. 98:602-604.
- Eberhart, S.A., M.N. Harrison and F. Ogata. 1967. A comprehensive breedng system. Der Zuchter 37:169-174.
- Fuller, J.P. 1934. Vegetative and reproductive responses associated with fruit development in cucumber,. Cornell Univ. Agr. Expt. Sta. Memo 163.
- Horst, E.K. and R.L. Lower. 1978. Cucumis hardwickii: A source of germplasm for the cucumber breeder. Cucurbit Genetics Coop. Rpt. 1:5.
- McCollum, J.P. 1934. Vegetative and reproductive responses associated with fruit development in cucumber. Cornell Univ. Agr. Expt. Sta. Memo 163.
- Nienhuis, J. and R.L. Lower. 1980. Influence of reciprocal donor scions on fruit setting characteristics of recipient scions of Cucumis sativus and C. hardwickii R. Cucurbit Genetics Coop. Rpt. 3:17-19.
- Rao, C.R. 1964. The use and interpretation of principal component analysis in applied research. Snakhya 26:329-357.
- Schuman, D.A., J.E. Staub and B.E. Sstruckmeyer. 1985. Morphological and anatomical comparisons between two Cucumis sativus botanical varieties: hardwickii and sativus. Cucurbit Genetics Coop. Rpt. 8:15-18.
- Smith,. O.S., R.L. Lowerand R.H. Moll. 1978. Estimates of heritabilities and variance components in pickling cucumner. J. Amer. Soc. Hort. Sci. 103:222-225.
- Staub, J.E. 1985. Preliminary yield evaluation of inbred lines derived from Cucumis sativus var. hardwickii (Royle) Kitamura. Cucurbit genetics Coop. Rpt. 8:18-21.
- Staub, J.R. and R.S.Kupper. 1985. Use of Cucumis sativus var. hardwickii germplasm in backcrosses with Cucumis sativus var. sativus. HortSci. 20:436-438.
- Tiedjens, V.A. 1928. Sex ratios in cucumber flowers as effected by different conditions of soil and light. J. Agr. Res. 36:721-746.