Cucurbit Genetics Cooperative Report 21:1-5 (article 1) 1998
Jinsheng Liu
Agricultural Division, Jiangsu State Farms Groiup Corporation, No. 1 Zhu Jiang Road, Nanjing, 210008, P. R. China
Jack E. Staub
Vegetable Crops Research Unit, USDA/ARS, Department of Horticulture, University of Wisconsin, Madison, WI 53706 U.S.A.
Introduction. Research using populations developed from diverse breeding lines indicates low genetic variance for fruit yield in processing cucumber (Cucumis sativus var. sativus L.) (Wehner, 1989). This low variance is at least particilly due to the narrow genetic base of pickling cucumber. The results of previous studies suggest that selection for yield in elite adapted populations would likely be a slow and costly process.
Incorporation of quantitatively inherited characteristics into commercially adapted cultivars from exotic germplasm can be an effective way to obtain greater genetic variation and response to selection (Wehner, 1989).Cucumis sativus var. hardwickii (R.) Alef. is a potential source of exotic germplasm for increasing the genetic variation for yield in cucumber because of its multiple lateral, sequential fruiting habit (Kupper and Staub, 1998, Frederick and Staub, 1989). Nevertheless, var. hardwickii has many undesirable characteristics which have caused problems during the incorporation of desirable characteristics. Other botanical varieties of Cucumis sativus exist and should be evaluated for their breeding potential.
The USDA North Central Plant Introduction Station (NCPIS), Ames, Iowa received 12 accessions which were classified as botanical varieties of C. sativus Table 1). These accessions by definition of the variety status were presumed to be different from C. sativus var. sativus, and were given the taxonomic classification of var. anatolicus (1; PI 504559), var. cilicicus (2; PI 504561), var. europaeus (3; PI 504562), var. falcatus (4; PI 504563, var. indo-europaeus (5; PI 504565), var. irano-turanieus (6; PI 504566), car. izmir (7; PI 504567), var. sikkimensis (8; PI 504568), var. squamosus (9; PI 504570), var. testudaceus (10; PI 504571), var. tuberculatus (11; PI 504572), and var. vulgatus (12; PI 504573) at their source prior to transmittal to NCPIS. Therefore, a study was designed to evaluate the genetic diversity among these accessions, and between these accessions and an accession of var. hardwickii (13; PI 183967) and var. sativus (Line GY 14) (14) used in previous studies (Kupper and Staub, 1998, Fredrick and Staub, 1989).
Material and Methods. Twelve accessions of putative botanical varieties of C. sativus were received from NCPIS (Table 1). These accessions along with C. sativus var. hardwickiiand C. Sativus var. sativus (line GY14) and F1 progeny (derived from crosses with these accessions and USDA WI 2870) were evaluated for isozyme variation at 21 loci according to Meglic et al. (1996) and five morphological characteristics [days to flower, lateral branch number (primary), cumulative fruit number, weight, and length:diameter ratio over three harvests]. For morphological analysis, varieties and F1 hybrids were planted in treatment rows 6.1 m long spaced 1/5 m apart at Hancock, WI in 1993. Plant spacing within a row was about 12 cm. Border rows were placed between main plots and at plot ends. For electrophoresis, accessions (~ 15 seedlings bulked) were characterized using starch gel electrophoresis. ANOVA and mean separation and cluster analyses were performed according to Steel and Torrie (1980). Mean separations for all variables in all ANOVAs were performed with Fisher’s protected LSD at the 5% level and coefficients of variation were calculated.
Results and discussion. Isozyme and morphological analysis resulted in similar, but not identical dendrogram depictions. Variation at nine polymorphic enzyme loci (Ak-2, Ak-3, Fdp-1, Mpi-2, Pgi Pgm, Pgd-2) was important in defining varietal differences (Table 2). Variety hardwickii and var. falcatus were most dissimilar from all other varieties examined (cluster analysis not presented).
Botanical varieties differed in their morphological characteristics (Table 1). Mean comparisons of morphological characteristics among the F1 progeny are given in Figure 1. Relative relationships based on morphology are depicted by cluster analysis in Figure 2. The following observations can be madeL 1) var. testudaceus was early flowering when compared to sativus; 2) var. sikkimensis produced more laterals than did sativus; 3) var. europaeus and vulgatus yielded higher than sativus4) var. sikkimensis had larger fruit than did all other varieties; and 5) var. falcatus produced longer fruit than did any otehr variety. The following observations can be made based on F1 performance: 1) var. hardwickii is probably the best source for increasing fruit number in variety sativus among the accessions examined in this study; 2) the varieties sikkimensis, vulgatus, and europaeus are similar but distinct from variety sativus which is most like cilicicus, imir and tuberculatusl and 3) since varieties sikkimensis and vulgatus produce an abundant number of lateralo branches and have genetic factors which condition relatively long and large fruit, these shold be considered as having potential for increasing yield potential in adapted cucumber. The variety europaeus also may have some potential in this regard. Varieties sikkimensis, falcatus, and hardwickii might have potential in plant improvement based on yield performance.
Table1. Comparison among putative Cucumis sativus L. botanical varieties for sex expression, days to anthesis, lateral branch number (primary), and fruit number, weight and length: diameter ratio (L:D).
|
Botanical variety
|
Origin
|
Sex1
|
Days to anthesis
|
Lateral branch number
|
Three harvest yeild
|
Three harvest weight (kg)
|
L:D ratio
|
|---|---|---|---|---|---|---|---|
|
var. anatolicus
|
Russia
|
M
|
36.8
|
2.5
|
58.8
|
36.1
|
4.0
|
|
var. cilicicus
|
Russia
|
M
|
19.0
|
1.0
|
81.5
|
29.5
|
1.9
|
|
var. europaeus
|
Russia
|
M
|
32.3
|
3.0
|
99.5
|
33.3
|
3.3
|
|
var. falcatus
|
Japan
|
M
|
44.5
|
1.5
|
30.0
|
18.6
|
11.6
|
|
var. indo-europaeus
|
Russia
|
M
|
35.3
|
2.8
|
86.5
|
31.8
|
2.1
|
|
var. irano-turanieus
|
Russia
|
M
|
35.0
|
2.8
|
82.3
|
27.5
|
2.1
|
|
var. izmir
|
Russia
|
M
|
35.8
|
1.5
|
93.5
|
29.2
|
2.1
|
|
var. sikkimensis
|
Russia
|
M
|
58.3
|
4.5
|
60.0
|
24.5
|
2.0
|
|
var. squamosus
|
Russia
|
M
|
38.0
|
2.8
|
81.8
|
17.5
|
1.8
|
|
var. testtudaceus
|
Russia
|
M
|
38.5
|
1.0
|
92.8
|
31.9
|
2.1
|
|
var. tubercullatus
|
China
|
M
|
35.0
|
0.0
|
45.0
|
19.4
|
3.2
|
|
var. vulgatus
|
India
|
M
|
39.5
|
3.3
|
48.3
|
23.6
|
2.4
|
|
var. hardwickii
|
India
|
M
|
77.5
|
6.0
|
2.0
|
0.2
|
1.5
|
|
var. sativus (Gy14)
|
USA
|
G
|
38.8
|
0.0
|
97.5
|
25.0
|
2.6
|
|
LSD
|
2.7
|
0.6
|
16.3
|
6.9
|
0.3
|
||
|
C.V. (%)
|
25
|
25
|
15
|
17
|
10
|
1M = monoecious and G = gynoecious
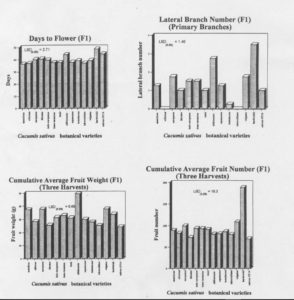
Figure 1. Comparisons among F1 hybrids derived from matings between putative botanical varieties of cucumber (Cucumis sativus L. )and USDA line WI 2870 (var. sativus) [Vertical bar 1 = var. anatolicus, 2 = var. cilicicus, 3 = var. europaeus, 4 = var. falcatus, 5 = var. indo-europaeus, 6 = var. irano-turanieus, 7 = var. izmir, 8 = var. sikkimensis, 9 = var. squamosus, 10 – var. testudaceus, 11= var. tuberculatus, 12 = var. vulgatus, 13 = var. hardwickii, and 14 = var. sativus (Gy14)].
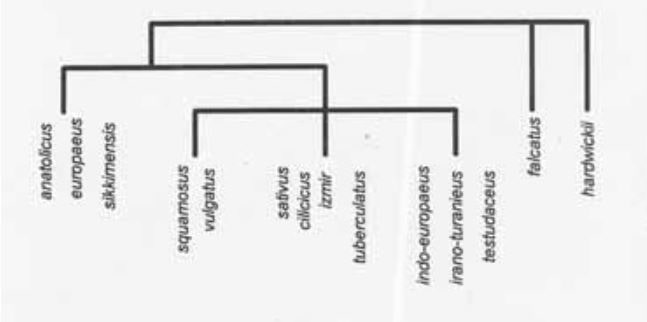
Figure 2. Relationships of cucumber (Cucumis sativus L.) accessions based on cluster analysis of morphological characteristics.
Table 2. Isozyme genotypes at nine loci of 14 Cucumis sativus botanical varieties1.
|
Botanical variety
|
Ak2
|
Ak3
|
Fdp1
|
Fdp2
|
Mdh3
|
Mpi2
|
Pgl
|
Pgm
|
Pgd2
|
|---|---|---|---|---|---|---|---|---|---|
|
sativus
|
22
|
11
|
22
|
11
|
11
|
22
|
22
|
11
|
11
|
|
anatolicus
|
12
|
22
|
22
|
11
|
11
|
22
|
22
|
12
|
22
|
|
cilicicus
|
22
|
12
|
12
|
11
|
11
|
11
|
22
|
12
|
12
|
|
europaeus
|
22
|
12
|
12
|
11
|
11
|
12
|
12
|
22
|
11
|
|
falcatus
|
22
|
11
|
11
|
11
|
11
|
11
|
22
|
22
|
11
|
|
hardwickii
|
11
|
11
|
12
|
11
|
11
|
22
|
22
|
22
|
11
|
|
indo-europaeus
|
22
|
22
|
11
|
22
|
11
|
11
|
11
|
12
|
22
|
|
irano-turanieus
|
22
|
22
|
22
|
22
|
12
|
11
|
11
|
22
|
22
|
|
izmir
|
22
|
22
|
12
|
22
|
11
|
22
|
11
|
12
|
22
|
|
sikkimensis
|
22
|
11
|
22
|
11
|
11
|
22
|
22
|
22
|
11
|
|
squamosus
|
12
|
22
|
11
|
22
|
11
|
12
|
22
|
22
|
22
|
|
testudaceus
|
22
|
11
|
22
|
11
|
11
|
11
|
11
|
11
|
11
|
|
tuberculatus
|
12
|
11
|
12
|
11
|
11
|
12
|
22
|
22
|
11
|
|
vulgatus
|
22
|
11
|
12
|
11
|
11
|
11
|
22
|
22
|
11
|
1only polymorphic loci shown
Literature Cited
- Fredrick L.R., and J.E. Staub. 1989. Combining ability analysis and evaluation of nearly homozygous lines derived from cucumis sativus var. hardwickii (R.) Alef. J. Amer. Soc. Hort. Sci. 114:332-338.
- Kupper, R.S., and J.E. Staub. 1988. Combining ability between lines of cucumis sativus L. and Cucumis sativus var.hardwickii (R.) Alef. Euphytica 38:197-120.
- Melgic, V., F. Serquen and J.E. Staub. 1996. Genetic diversity in cucumber (Cucumis sativus L.): I. A reevaluation of the U.S. Germplasm collection. Genet. Res. Crop. Evol. 46:533-46.
- Steel, R. and J.H. Torrie. 1980. Principles and procedures of statistics. 2nd ed. McGraw-Hill, New York.
- Wehner, T.C. 1989. Breeding for improved yield in cucumber, p. 323-359. In: J.Janik (ed.), Plant breeding reviews, vol. 6. Wiley, New York.