Cucurbit Genetics Cooperative Report 23:4-7 (article 2) 2000
G. Fazio and J.E. Staub
USDA-ARS Vegetable Crops Research Unit, University of Wisconsin-Madison, Department of Horticulture, 1575 Linden Dr., Madison WI 53706
Introduction. Genetic markers such as isozymes, RAPDs and RFLPs have ben characterized in cucumber (Cucumis sativus L.) (Dijkhuizen et al., 1996; Serquen et al., 1997). However, critical documentation of these markers and their usefulness in marker-assisted selection (MAS) for applied breeding programs has been limited. This is at least partially due to the species’ low polymorphism level. A higher level of polymorphism has been associated with SSR loci in preliminary studies with C. sativus well as in other genera (Cucurbita and Citrullus) of the Cucurbitaceae (Katzir et al., 1996). In this study 12 of 20 SSRs (60%) possessed two to give alleles per locus.
A moderately large molecular marker database now exists for cucumber (Meglic et al., 1996a and 1996b; Dijkhuizen et a;., 1996). The number of polymorphic loci identified in these crops is useful for variety identification and seed purity testing, but is not robust enough to be used for legal applications or some phases of germplasm management (Staub et al., 1999). The identification and characterization of SSR markers would allow for more precision in the estimation of genetic similarities among cucumber cultivars, and thus provide cucumber researchers with additional markers for genetic analysis (Staub et al., 1996). Since their initial implementation, PCR markers based on microsatellites have become the marker of choice for many genetic. Their codominant nature, and high polymorphism rate, and high through-put capacity are important methodological characteristics. The initial development of microsatellite markers requires the characterization of sequences flanking the repeat motif, followed by the design of flanking PCR primers. Many methods have been devised for the initial characterization of the flanking sequences (Litt and Luty, 1989). Other methods include: database search, construction of a genomic or cDNA library and screening with oligonucleotide probes, and construction of microsatellite-enriched libraries followed by PCR screening. We describe herein a novel method to capture sequences that contain microsatellite motifs.
Materials and Methods:DNA extraction, restriction and size fractionation. DNA from cucumber breeding line G421 was extracted from young leaves and apical meristems according to the nuclear DNA extraction protocol outlined in Maniatis (1982). Extraction products were treated with RNAse ONE (Promega, Madison, WI). DNA was quantified using a TKO 100 fluorometer (Hoefer). In separate reactions 100 mg of DNA were restricted using EcoR I restriction enzyme (Promega, Madison WI) and Acs I (Boehringer Mannheim) which recognizes the sequence (A or G)/AATT(T or C) and generates compatible ends with EcoR I. Restricted DNA was size fractionated using a low pressure 75cm long 16mm diameter chromatography column (Fisher) S-500 (Pharmacia). The column was packed according to instructions for Sephacryl S-500 and equilibrated with 200 ml elution buffer at 0.3 ml/min (0.1 M tris-HCL pH 8.0, 0.15 M NaCl, 0.001 M ETDA). Both samples of restricted DNA were fractionated separately and eluted DNA was collected at 2 ml aliquots. Eluted DNA wa precipitated, washed and resuspended in TE. Elution aliquots were sized by agarose gel electrophoresis. Fractions between 200 bp to 1200 bp were combined for ligation to vector.
Library construction, mass excision and plasmid DNA extraction. Fractionated DNA was ligated to Ziplox EcoR I arms (Life Technologies, Gaithersburg, MD) and packaged in a lambda vector with Gigapack III Gold packaging extract (Stratagene, La Jolla, CA). The resulting library was titered and efficiency of packaging was determined by blue/white colony screening. Primary Ziplox libraries were max excised in vivo into the pZL1 plasmid vector (Life Technologies, Gaithersburg, MD) in DG10B Zip (Life Technologies, Gaithersburg, MD) strain of E. coli. The mass excision and expansion of the plasmid library were performed at the same time in a semisolid media to minimize representational biases that can occur during expansion in liquid media. Plasmid DNA was extracted b7 a plasmid maxi-prep procedure (Quiagen, Valencia, CA).
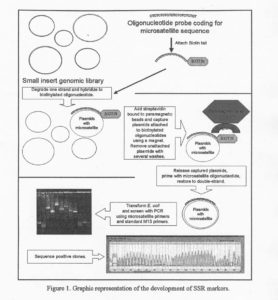
Capture of plasmids containing microsatellites. In preparation for the GeneTrapperTM (Life Technologies, Gaithersburg, MD) reaction, several oligonucleotides (20-30 bp) coded for common microsatellite sequences (CT, TG, CTT, TCC, ATT) were biotinylated (biotin-14-dCTP) with a terminal transferase enzyme (Life Technologies, Gaithersburg, MD). The “capture” steps are described in detail in the GeneTrapper kit (Life Technologies, Gaithersburg, MD) and graphically represented in Figure 1.
- The plasmid DNA was treated with Gene II protein and Exonuclease III to obtain single stranded circular plasmid DNA.
- Single stranded DNA was hybridized with the biotinylated oligonucleotides. During the hybridization, the single stranded oligonucleotides hydrogen bonded with their complementary microsatellite sequences.
- Para-magnetic beads bound to streptavidin were used to capture the microsatellite containing single stranded plasmid molecules. The product of the capture was washed several times to eliminate unbound single stranded DNA.
- The final product of the capture was then treated with a buffer to release the captured single stranded DNA. The single stranded DNA was subsequently primed and repaired to form double-strand DNA.
- The double-stranded plasmids were then used to transform DH10B E. coli that had been grown on ampicillin selective media. It was assumed that only E. coli harboring the intact pZL1 (putatively containing a microsatellite region) coming from the cucumber DNA library survived the selection media.
- Single colonies were picked, named sequentially, and then amplified prior to plasmid DNA extraction. Confirmation of the presence of a microsatellite region was obtained by subjecting plasmid DNA to PCR using the standard forward or reverse M13 and a primer coding for the captured microsatellite type.
- Plasmid DNA from positive clones was sequenced at the University of Wisconsin Biotechnology Center to identify the microsatellite type and length.
Sequence analysis and primer design. All sequences from positive clones were analyzed and compared to each other using the assembly feature in the GeneTool software package (Biotools, Edmonton, Canada) in order to discard duplicates and identify overlapping DNA regions. The resulting unique sequences were entered into the primer design software OLIGO 6.0 (Life Science Software, Long Lake, MN).
Results and discussion: The ease of application of the Gene TrapperTM System (Life Technologies) made it possible to isolate and characterize several cucumber microsatellite loci in a relatively short time without the use of radiolabeled products. These results were obtained in 1998 independently of a similar technique developed by Paetkau (1999).
This methodology has been repeated five times with success and reproducibility of several types of microsatellites (di-, tri-, and tetra-nucleotides). Nevertheless, this methodology continues to be further refined and is still considered to be in its developmental stages. The first capture was performed with a (CT) 12 oligonucleotide, and the procedure yielded 457 clones. The PCR assays identified the size of the insert and its relative position. Based on these assays, 300 of the 457 clones were selected for the primer design step of the SSR construction. The insert sequences were entered into the Oligo 6.0 primer design tool (Molecular Biology Insights Inc., Long Lake, MN).
Results and Discussion: The ease of application of the GeneTrapperTM System (Life Technologies) made it possible to isolate and characterize several cucumber microsatellite loci in a relatively short time without the use of radiolabeled products. These results were obtained in 1998 independently of a similar technique developed by Paetkau (1999).
This methodology has been repeated five time with success and reproducibility of several types of microsatellites (di-, tri-, and tetra-nucleotides). Nevertheless, this methodology continues to be further refined and is sill considered to be in its developmental stages. the first capture was performed with a (CT) 12 oligonucleotide, and the procedure yielded 457 clones. The PCR assays identified the size of the insert and its relative position. Based on these assays, 300 of the 457 clones were discarded as suspected duplicates. Sixty-five clones were selected for the primer design step of the SSR construction. the insert sequences were entered into the Oligo 6.0 primer design tool (Molecular Biology Insights Inc., Long Lake, MN).
Other captures using (TG)12, (CTT)8, (TCC)8, (ACTC)6, (ATT)9 have been successfully performed, and are currently being characterized as with the CT repeat capture. All primer pairs that have been designed are being tested on C. sativus lines G421 and H-19 using a thermal gradient PCR programmed on an Eppendorf Mastercycler Gradient thermal cycler (Hamburg, Germany) to establish the optimum annealing temperature range for each SSR primer.
This information will be used later in the development of PCR procedures for multiplexing SSR markers/ The SSR markers being developed in cucumber are being tested on C. melo parent lines MMR-1 and ‘Topmark’ to establish the extent of their cross-compatibility and potential use in melon. To date, approximately 80% of the SSR markers that have been constructed have amplified in both C. sativus and C. melo. the development of SSR markers will be useful for mapping, and introgressing economically important traits into cultivated varieties.
Literature Cited
- Dijkhuizen, A., W.C. Kennard, M.J. Havey, and J.E. Staub. 1996. RFLP variation and genetic relationships in cultivated cucumber. Euphytica 90: 79-87.
- Katzir N., Danin-poleg Y., Tzuri G., Karchi Z., Lavi U., and Cregan P.B. 1996. Length polymorphism and homologies of microsatellites in several cucurbitaceae species. Theor. Appl. Genet. 93: 1282-1290.
- Litt, M. and J.A. Lutty. 1989. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle acting gene. Am. J. Hum. Genet. 44:397-401.
- Maniatis, T., E.F. Fritsch, and J. Sambrook. 1982. Molecular Cloning: A Laboratory Manual. cold Spring Harbor publisher, Cold Spring Harbor, NY.
- Meglic, V., and J.E. Staub. 1996a. Genetic diversity in cucumber (Cucumis sativus L) II. An evaluation of selected cultivars released between 1846 and 1978. Genet. Res. Crop. Evol. 43:547-558.
- Meglic, V., and J.E. Staub. 1996b. Inheritance and linkage relationships of isozyme and morphological loci in cucumber (Cucumis sativus L). Theor. Appl. Genet. 92: 865=872.
- Paetkau, D. 1999. Microsatellites obtained using strand extension: an enrichment protocol. Biotechniques 26:690-697.
- Serquen, F.C., J. Bacher and J.E. Staub. 1997. Mapping and QTL analysis of horticultural traits in a narrow cross in cucumber (Cucumis sativus L.) using random-amplified polymorphic DNA markers. Mol. Breed. 3:257-268.
- Staub, J.E., F. Serquen, and M. Gupta. 1996. Genetic markers, map construction, and their application in plant breeding. HortScience 31:729-741.
- Staub, J.E. 1999. Intellectual property rights, genetic markers, and hybrid seed production. J.New Seeds 1:39-65.