Cucurbit Genetics Cooperative Report 8:22-25 (Article 9) 1985
Staub, J. and L. Frederick
U. S. Department of Agriculture, Agricultural Research Service and Department of Horticulture, University of Wisconsin, Madison, WI 53706
Previously (9), we have documented the relative activity of 47 metabolic enzymes, and general protein in cucumber (Cucumis sativus L.). In conjunction with this study we determined that enzyme polymorphisms existed in glucosephosphate isomerase (GPI), glutathione reductase (GR), isocitrate dehydrogenase (IDH), peptidase with phenyl-alanyl-proline (PEP-PAP) and phosphoglucomutase (PGM). We used additional enzymes (16) in order to obtain an initial estimate of the potential genetic difference between several species in the anguria and sativus groups (8) as classified by Esquinas-Alcazar (4).
Preliminary data from this study indicated that, although species within the groups evaluated share some common banding patterns, enough difference existed between them to suggest that their genetic distance is certainly as great as Esquinas-Alcazar stated. The objective of this study was to survey the electrophoretic variability within and between cross-compatible and cross-incompatible wild Cucumis species in order to: 1) document the relative mobility of electromorphs observed in 7 enzyme systems and; 2) provide information which might lead to a better understanding of the biosystematics of this genus.
Cotyledonary extracts of 8 wild Cucumis species and 1 Cucumis sativus L. inbred processing cucumber line (Gy 14A) were examined by horizontal starch (12%) gel electrophoresis. The plant introductions examined included accessions of 4 annual (C. africanus Lindley F., C. anguria L., C. dipsaceus Spach and C. myriocarpus Naud.), and 2 perennial (C. ficifolius A. Rich and C. zeyheri Sond.) cross-compatible wild diploid (2n=24) species. The autotetraploid, C. heptadactylus Naud. (2n=48), was also examined since it is considered a member of this cross-compatible group based on hybridization studies by Deakin et al. (3).
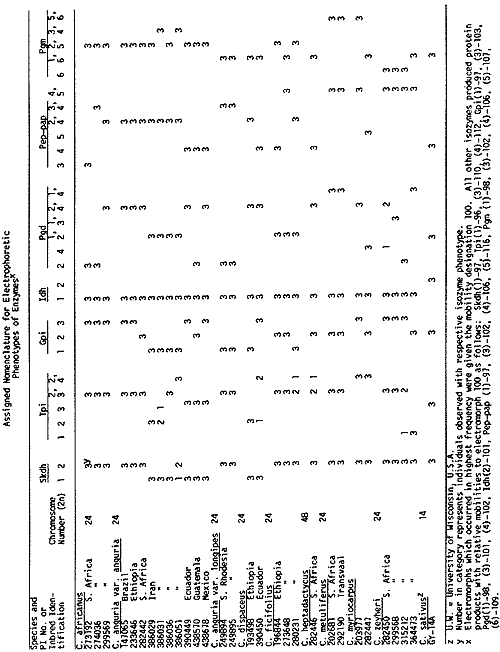
Isozyme banding patterns of shikimic dehydrogenase (SKDH), triose phosphate isomerase (TPI), GPI, IDH, PGD, PEP-PAP, and PGM were recorded and comparisons were made among zymograms (Table 1). In order to standardize the relative mobilities of the observed electromorphs, extracts of Gy 14 were loaded on each gel, and histochemical staining for specific enzymes was performed according to Shaw and Prasad (6) and Allendorf et al. (1).
The nomenclature follows a modified form described by Richmond (5) such that isozymes for the enzymes SKDH, TPI, GPI, IDH, PGD, PEP-PAP and PGM are designated as Skdh, Tpi, Gpi, Idh, Pgd, Pep-pap and Pgm, respectively. Numerals refer to isozymes numbered from the most cathodal to the most anodal region of the gel. For each enzyme, the most common allele was designated as 100. As an example, the combination of homomeric protein products of the 2 singlebanded electromorphs, 2 (100) and 3 (110), at the Tpi locus (which has at least 4 single-banded electromorphs), produce a heteromeric product which is designated Tpi 2,3 with the relative isozyme mobilities of 100/110.
Electrophoretic variation was observed within and between cross-compatible and cross-incompatible groups for all enzyme systems with the exception of IDH which was monomorphic. All the species examined are apparently fixed for Idh 1 (100), while Gy 14 possesses Idh 2 (101) which appears to be characteristic of C. sativus var. sativus (7). Variation for SKDH exists within C. anguria var. anguria which possesses both Skdh 1 and 2, while all other species exhibit Skdh 2 except for C. dipsaceus which produced only Skdh 1 electromorphs. All species appeared to possess Tpi 2 (100) and/or 3 (102) except for . anguria var. anguria PI 386051, C. dipsaceus PI 390450, C. ficifolius PI 280231, C. heptadactylus PI 282446 and C. myriocarpus PI 203977 and PI 282447 which produced the electromorph 4 (112).
Tpi 1 (96) was found exclusively in C. zeyheri PI 315212 and PI 364473. Gpi staining produced 3 single-banded electromorphs for which C. africanus was monomorphic for Gpi 3, C. metuliferus for Gpi 2 and C. anguria var. longipes for Gpi 1. While C. anguria var. anguria exhibited the isozyme phenotypes Gpi 1, 2 and 3, the other species segregated 1 and 3 (C. dipsaceus), 1 and 2 (C. ficifolius) and 2 and 3 (C. zeyheri and C. myriocarpus). C. dipsaceus, C. metuliferus and C. ficifolius were monomorphic for Pgd 2,4, Pgd 1,4 and Pgd 1,2, respectively. Only C. zeyheri PI 299568 showed evidence of Pgd 3 (101). Evidence for Pep-pap 3 (102) was exclusively observed in C. africanus and C. anguria var. longipes, indicating a closer relationship than had previously been thought. Likewise, evidence for Pep-pap 5 (116) was uniquely recorded in C. metuliferus, C. myriocarpus, C. ficifolius and C. zeyheri, suggesting a comparatively close relationship between these species. Evidence for Pgm 5 (107) was observed in C. africanus, C. anguria var. anguria, C. ficifolius, C. myriocarpus and C. metuliferus. In contrast, C. metuliferus and C. myriocarpus possessed Pgm 6 (109) which was also recorded in all species except C. africanus, and C. anguria var. anguria. On the other hand, C. metuliferus and C. myriocarpus exhibited for Pgm 5 (107) which was also present in C. africanus and C. anguria var. anguria.
It appears from these preliminary data that the ancestry of C. metuliferus and C. myriocarpus may be relatively close. It is also interesting to note that several C. anguria var. anguria accessions from Iran showed isozyme variations which set them apart from the rest of the var. anguria collections. Moreover, var. longipes and var. anguria are different for PEP- PAP and PGM, confirming (2) their varietal difference.
Table 1. Electrophoretic Variation Observed for Shikimic dehydrogenase (SKDH), triose phosphate isomerase (TPI), glutamic pyruvic transminase, glucosephosphate isomerase (GPI), phosphogluconate dehydrogenase (PGD), peptidase with phenyl-alanyl-proline (PEP-PAP) and phosphoglucomutase (PGM) in 8 Cucumis species.
Literature Cited
- Allendorf, F. W., N. Mitchell, N. Ryman, and G. Stahl. 1977. Isozyme Loci
in Brown Trout (Salmo trutta L.): Detection and Interpretation from
Population Data. Hereditas 86:179-190. - Dane, F., D. W. Denna and T. Tsuchiya. 1980. Evolutionary studies of wild
species in the genus Cucumis. Z. Pflanzenauchtg. 85:89-109. - Deakin, J. R., G. W. Bohn, and T. W. Whitaker. 1971. Interspecific
Hybridization in Cucumis. Econ. Bot. 25:195-211. - Esquinas-Alcazar, J. T. 1977. Alloenzyme Variation and Relationships in
the Genus Cucumis. Ph.D. Thesis, University of California, Davis,
California. - Richmond, R. C. 1972. Enzyme variability in the Drosophila williston Group
3. Amounts of Variability in the Superspecies D. paulistorum. Genetics
70:87-112. - Shaw, C. R. and R. Prasad. 1970. Starch Gel Electrophoresis of Enzymes —
A compilation of Recipes. Biochem. Genet. 4:297-320. - Staub, J. E., R. S. Kupper, D. Schuman, T. C. Wehner and B. May. 1985.
Electrophoretic Variation and Enzyme Storage Stability in Cucumber. J.
Amer. Soc. Hort. Sci. (In press). - Staub, J. E., R. S. Kupper and T. C. Wehner. 1984. Electrophoretic
Comparison of Six Species of Cucumis. Cucurbit Genetics Coop. Rpt.
6:27-30. - Staub, J. E., R. S. Kupper, and T. C. Wehner. 1983. Preliminary Evaluation
of Isozyme Polymorphisms in Cucumis. Cucurbit Genetics Coop. Rpt. 6:32-
34.