Cucurbit Genetics Cooperative Report 8:26-28 (Article 10) 1985
Strefeler, Mark S. and Todd C. Wehner
Department of Horticultural Science, North Carolina State University Raleigh, NC 27695-7609
An effective plant breeding program depends on the existence of genetic variability, and the estimation of genetic variance and gene effects are vital in formulating the most effective breeding procedures for the traits in question. Comstock and Robinson (1) provided several methods for obtaining information and estimates of variability and gene effects in plant populations. These provide the breeder with useful information about the breeding populations available to him. This study was undertaken because no estimates of genetic variance components are available for fresh-market cucumbers (Cucumis sativus L.) or for cucumber populations varying in degree of genetic diversity. The objectives of this study were to obtain estimates of the genetic variance components for fruit yield and quality traits in 3 freshmarket cucumber populations, and to predict gains from selection for each population based on the above estimates.
Methods. Three North Carolina fresh-market (slicer) cucumber populations were used for this experiment: NC Elite Slicer 1 (NCES1), NC Medium Base Slicer (NCMBS), and NC Wide Base Slicer (NCWBS). The 3 populations were developed with increasing genetic diversity. A North Carolina Design I was used to estimate the variance components in the 3 reference populations. The construction and analysis of this design was given by Comstock and Robinson (1). In this study, 72 SO plants from each population were chosen at random and designated as males. Each male was mated with 3 SO plants chosen at random and designated as females. In order to estimate genotype x environment interactions, 2 environments were used in 1984 — spring and summer. The seeds produced by the mating design were planted at the Horticultural Crops Research Station near Clinton, NC using a nested design with full-sib families (females) nested in half-sib families (males). Fruit were harvested at the green stage as described by Wehner and Miller (4) for optimum efficiency in measuring yield in a once-over harvest. Plots were defoliated using Paraquat 1,1′-dimethyl-4,4′-bipyridinium ion) as recommended by Wehner et. al. (5) to make data collection at harvest stage more efficient. Total and marketable (total fruit number – number of culls) fruit yield per plot were estimated by counting as recommended by Ells and McSay (2). Early yield was measured by counting the number of oversized fruit (<50mm dia.) per plot. Fruit shape, seedcell size, and overall impression were rated on a scale of 1 to 9 with 1=poor, 5=average, and 9=excellent. Color was rated on a scale of 1 to 9 with 1=white, 5=light green, and 9=very dark green.
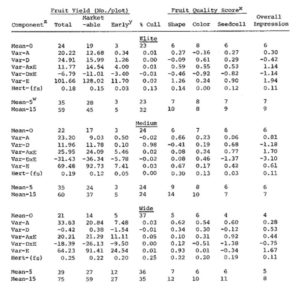
For each of the 3 populations, 9 variables were calculated as described by Hallauer and Miranda (3) (Table 1). These were the mean (Mean-O), additive genetic variance (Var-A), dominance variance (Var-D), additive by environmental variance (Var-AxE), dominance by environmental variance (Var- DxE), environmental variance (Var-E), heritability of half-sib selection (Hert.-hs), mean after 5 selection cycles (Mean-5), and mean after 15 selection cycles (Mean-15).
Results. Means for the 4 yield and 4 fruit quality traits differed, as expected, among populations with the elite population having the highest means and the wide base population having the lowest. The one exception was for early yield where the wide base population was the highest and the elite and medium base populations were equal. Additive variance was predominant for shape and overall impression in the elite and medium base populations and for seedcell size and early yield in the wide base population. Dominance variance predominated for color in the elite population, for percent culls in the medium base population, and for total yield and marketable yield in all populations. Heritabilities were low to moderate (0.00-0.23) in the 3 populations for all traits studied. Heritabilities of the wide base population were generally higher than heritabilities observed in the elite and medium base populations, except for overall impression which was higher in the elite and medium base populations, and shape which was much higher in the medium base population than the other populations.
After 5 cycles of full-sib family selection, the predicted means for total yield were highest in the wide base population, but the elite population had the highest predicted means for all other traits except shape. After 15 cycles of full-sib family selection, predicted means for total, marketable, and early yield were 59,45, and 5 fruits/plot for the elite population, respectively; 60,37, and 5 fruits/plot for the medium base population, respectively; and, 75, 59, and 27 fruits/plot for the wide base population, respectively. Predicted means for fruit quality ratings ranged from 6 to 10 for the elite population, 6 to 14 for the medium base population, and 7 to 12 for the wide base population after 15 cycles of selection. The wide base population means surpass the elite and medium base population means after 15 selection cycles for all traits except percent culls, shape and overall impression. The low gain per selection cycle for overall impression in the wide base population may be caused by the large number of families with very low quality ratings which may have caused a downward bias in the estimate of additive variance in this population.
The wide base population is the best population for improving the traits measured in this study because it had the highest predicted means for all traits except percent culls, shape, and overall impression; and although the predicted means for overall impression and shape were not the highest, they were at an acceptable level (a mean score of 8) after 15 cycles of selection.
Table 1. Means, genetic variances, heritabilities and predicted gains for 4
yield and 4 quality traits studied in 3 cucumber populations (elite,
medium and wide) differing in genetic diversity.
zVar-A is the additive variance, Var-D is the dominance variance, Var-AxE is the additive by environmental variance, Var-DxE is the dominance by environmental variance, and Hert-(hs) is the heritability based on half-sib family means.
yEarly yield is the number of oversized fruits (<50 mm dia.) per plot at harvest.
xScored 1 to 9 (1=poor, 5=average, 9=excellent; except color which was scored 1=white, 5=light green, 9=very dark green). Scores above 9 indicate improvement in fruit quality traits to levels not now observed.
wMean-0, -5, -15 is the population mean after 0,5, and 15 cycles of selection, respectively.
Literature Cited
- Comstock, R. E. and H. F. Robinson. 1948. The components of genetic
variance in populations. Biometrics 4:254-266. - Ells, J. E. and A. E. McSay. 1981. Yield comparisons of pickling cucumber cultivar trials for once-over harvesting. HortScience 16:187-189.
- Hallauer, A. R. and J. B. Miranda Fo. 1981. Quantitative genetics in maize breeding. Iowa State University Press, Ames.
- Wehner, T. C. and C. H. Miller. 1984. Efficiency of single-harvest methods for measurement of yield in fresh-market cucumbers. J. Amer. Soc. Hort. Sci. 109:659-664.
- Wehner, T. C., T. J. Monaco, and A. R. Bonanno. 1984. Chemical defoliation of cucumber vines for simulation of once-over harvest in small plot yield trials HortScience 19:671-673.