Cucurbit Genetics Cooperative Report 17:116-119 (article 34) 1994
X.P. Zhang and B.B. Rhodes; H. Skorupska
Dept. of Horticulture, Clemson University, Clemson, SC 29634; Dept. of Agronomy and Soils and Biological Sciences, Clemson University, Clemson, Sc 29634
Watermelon is grown worldwide, yet genetic evaluation is far behind other important crops such as cucumber and melon (Henderson, 1991; Robinson, et al., 1976). More genetic information is needed for efficient genetic improvement of this crop.
Genetic markers closely linked with genes of interest are useful for efficient selection of the traits controlled by the genes in the breeding population. Navot and Zamir (1986) have developed a partial linkage map using isozyme and seed protein markers. Seven linkage groups were identified based on populations derived from interspecific crosses. Five of these groups consist of two or three marker loci (Navot and Zamir, 1986; Navot et al., 1990).
A major limitation of using Isozyme/protein markers is that limited polymorphic loci can be detected in watermelon, especially in the species C. lanatus (Bilew et al., 1989; Navot and Zamir, 1987. Zamir et al., 1984; Zhang and Wang, 1989. A new marker system, which offers a large number of polymorphic loci, is needed to develop a genetic map with high density or map the genomic regions around the genes of interest. DNA markers have introduced a new dimension to the development of genetic maps and the mapping of agronomically and physiologically important characters since they have the potential to reveal an almost unlimited number of polymorphisms. DNA markers commonly used are restriction fragment length polymorphisms (RFLPs) and random amplified polymorphic DNA (RAPD). RFLP assay detects DNA polymorphism through end nuclease digestion, coupled with DNA blot hybridization, and are in general time-consuming and labor-intensive. RAPD assay is based on the amplification of genomic DNA with single primers of arbitrary nucleotide sequence. These primers detect polymorphisms in the absence of specific nucleotide sequence information and the polymorphisms function as genetic markers. The major advantage of this assay is that there is no requirement for DNA sequence information. The protocol is relatively quick and easy to perform and automation is feasible. The RAPD assay uses fluorescence in lieu of radioactivity and only nanogram quantities of DNA are required. The RAPD markers have been used for many genetic studies of plant crops, and are suitable for marker assisted selection in applied breeding (Martin et al., 1991; Michelmore et al., 1991; Paran et al., 1991; Williams et al., 1991).
The objective of this study was to investigate RAPD molecular polymorphisms, and identify polymorphic RAPD markers for developing a molecular linkage map and mapping Fusarium wilt resistance gene(s) in watermelon.
Materials and Methods. We used three cultigens and one primitive watermelon for most of the primers tested: 1) 617AB (ms ms) , a male-sterile line with round fruit, red flesh and stripe rind; 2) ‘Dixilee’, a late variety with round fruit, red flesh and stripe rind; 3) PI 296341, a primitive watermelon with resistances to race ), 1 and 2 of the Fusarium wilt pathogen, and small round fruit with white flesh; and 4) ‘New Hampshire Midget’ (NHM), an early variety with small round fruit, red flesh, light green fruit color and susceptibility to all races of Fusarium wilt pathogens. Eight more genotypes and the F1 hybrid NHM x PI 296341 were used for testing some primers.
DNA Isolation and PCR Amplification. The DNA isolation procedures and PCR amplification conditions were the same as described by us (Zhang et al., 1992). Amplification was performed in a Perkins-Elmer/Cetus Model 480 DNA thermal cycler. Fifty-three 10-mer primers from the Operon RAPD Primer Kit C, E and M (Operon, Alameda, CA) were used in this study.
Cluster Analysis. A cluster analysis of four genotypes was performed using UPGMA (unweighted pair group method using arithmetic average) method on SAS program. Eighty-nine polymorphic RAPD loci observed from the four genotypes were used for the average cluster analysis.
Results. Three (5.6%) primers of the 53 primers tested were unable to prime amplification for the four watermelon genotypes under the conditions used in this experiment. By changing the reaction condition, amplification can be obtained from those primers that were unable to prime amplification in this experiment. Fourteen (26.4%) primers were only able to prime amplification for some genotypes. A total of 159 readable bands were obtained from the 36 primers that were able to prime DNA amplification for all the accessions tested, yielding 4.4 bands per primer. Among the total of 159 bands, 89 (56.0%) of the bands were polymorphic among the four accessions, 82 (51.6%) of the bands were polymorphic between NHM and PI 296341, but only 16 (10.1%) of the bands were polymorphic among the three cultigens. About 2.5 polymorphic bands were revealed per primer among the accessions tested. Therefore, RAPD markers offer a high level of polymorphism in watermelon when a primitive type was included. Fig. 1 shows the RAPD bands generated from cultigens 617AB (ms ms), ‘Dixilee’ and NHM, and primitive watermelon PI 296341 using primers OPM01, OPM01, OPM03, OPM04 and OPM05. Fig. 2 shows polymorphic RAPD loci generated from 13 watermelon genotypes using primer OPC08.
RAPD markers were further tested on F1 hybrid NHM x PI 296341 and its parents (NHM and PI 296341) to confirm polymorphism and inheritance of the RAPD marker. Similar to the RAPD marker detected in other crops (Williams et al., 1990; Michelmore et al., 1991) RAPD markers were inherited in a dominant fashion in watermelon (Fig. 2).
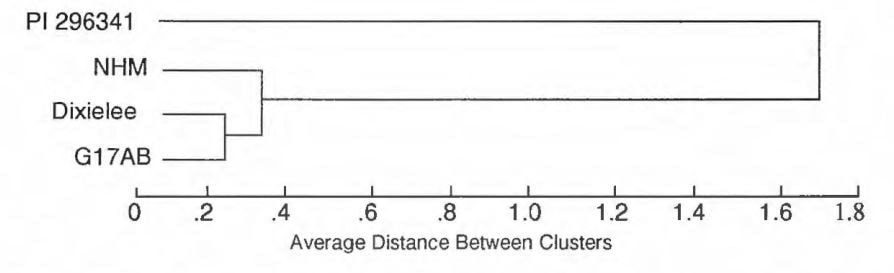
We did a cluster analysis using the RAPD data obtained from the three cultigens 61 7AB(ms ms), ‘Dixilee’ and NHM. and one primitive type PI 296341 to test if the RAPD data are correlated to the variation in agronomic/morphologic character. As shown in Fig. 3, 617AB(ms ms) and ‘Dixilee’, which are morphologically and agronomically similar, were joined as one cluster at an average distance of 0.239619, NHM was joined to the first cluster at an average distance of 0.262833, and PI 296341. which is distant from all three cultigens, was joined to the second cluster at an average distance of 1.744839. We confirmed the great genetic distance between cultigens and primitive watermelon at the DNA level. However, the three cultigens were very similar at the DNA level although they were developed at different times and locations. Breeding has considerably narrowed the genetic background of this crop.
In summary, the use of RAPD markers is an effected marker system for watermelon genome mapping because of the high polymorphism in the species. The existence of a relatively small genome (Arumuganathan and Earle, 1991; and our unpublished data) and a large number of polymorphic RAPD markers provides a potential for the development of a high density map efficiently. To be successful, however, a primitive type should be used as one of the parents to generate enough segregating marker loci in F2 or any population used for cosegregation analysis.
Figure 1 RAPD polymorphism in watermelon obtained with primers OPM – 01, OPM- 02, OPM-03, OPM-04, OPM-05. 1) PI 296341, 2) NHM, 3) G 17AB(msms), 4) Dixielee.
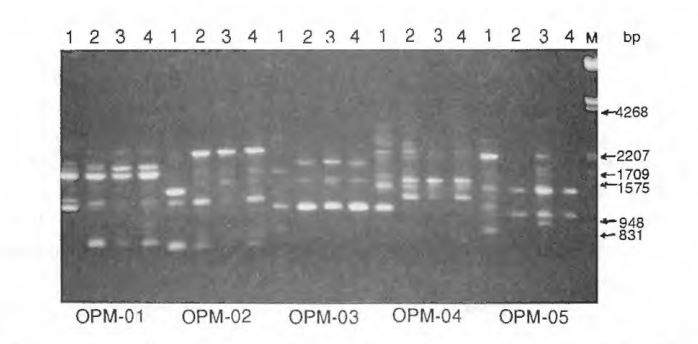
Figure 2 RAPD polymorphism in watermelon obtained with primer OPC- 08. 1) G 17AB(msms), 2) DiHielee, 3) PI 296341 , 4) NHM, 5) Tomato Seed Watermelon( tsts), 6) Edible Seed Watermelon, 7) SC-7, 8) bl-91, 9) NHMHP I 296341, 1 0) Au-Producer, 11) Sweet Princess, 12) Yellow Sweet, 13) G 17AB(Msms)
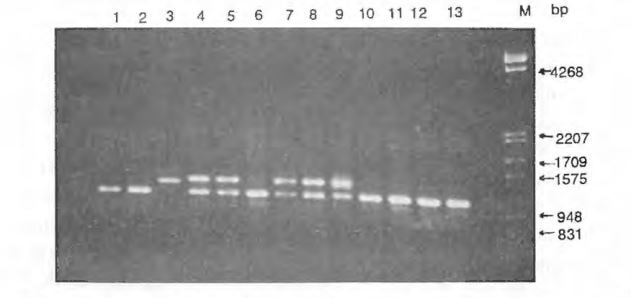
Figure 3. Average linkage cluster analysis of four watermelon genotypes based on 89 polymorphic RAPD markers.
Literature Cited
- Arumuganathan, K., and E.D. Earle. 1991. Nuclear DNA content of some important plant species. Plant Mol. Biol. Rpt. 9:208-218.
- Biles, C.L., R.D. Martyn, and H.D. Wilson. 1989. Isozymes and general proteins from various watermelon cultivars and tissue types. HortScience 24:810-812.
- Henderson, W., 1991. Gene list for watermelon. Cucurbit Genetic Cooperative Rpt. 14:129-137.
- Martin, G.B., J.G.K. Williams, and S.D. Tanksley. 1991. Rapid identification of markers linked to Pseudomonas resistance gene in tomato by using random primers and near isogenic lines. Proc. Natl. Acad. Sci. USA 88:2336-2340.
- Michelmore, R.W., I. Paran, and R.V. Kesseli. 1991. Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions using segregating populations. Proc. Natl. Acad. Sci. USA 88:9828-9832.
- Navot. N, and D. Zamir. 1986. Linkage relationships of 19 protein coding genes in watermelon. Theor. Appl. Genet. 72:274-278.
- Navot, N. and D. Zamir. 1987. Isozyme and seed protein phylogeny of the genus Citrullus (Cucurbitaceae). Plant Syst. Evol. 156-61-67.
- Navot, N., M. Sarfattio, and D. Zamir. 1990. Linkage relationships of genes affecting bitterness and flesh color in watermelon. J. Hered. 81:162-165.
- Paran, I., R. Kesseli, and R. Michelmore. 1991. Identification of restriction-fragment – length-polymorphisms and random amplified polymorphic DNA markers linked to downy mildew resistance genes in lettuce, using near isogenic lines. Genome 34: 1021-1027.
- Robinson, R.W., H.M. Munger, T.W. Whitaker, and G.W. Bohn. 1976. Genes of the Cucurbitaceae. HortScience 11:554-569.
- Williams, J.G.K., A.R. Kubelik, K.J. Livak, J.R. Rafalski, and S.V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535.
- Zamir, D., N. Navot and J. Rudich. 1984. Enzyme polymorphism in Citrullus lanatus and C. colocynthis in Israel and Sinai. Plant Syst. Evol. 146:163-170.
- Zhang, X.P., and M. Wang. 1989. Isozyme analysis of F1 hybrids and their parents in watermelon (Citrullus lanatus). Fruit Sci. 6(2):97-102 (in Chinese).
- Zhang, X.P., D. Heckel, L.C. Gahan, and B. Rhodes. 1992. Watermelon DNA isolation and amplification using the polymerase chain reaction. Cucurbit Genetics Cooperative Rpt. 15:87-89.